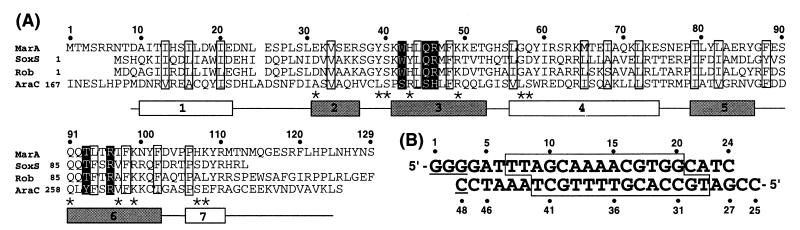
Figure 1.
Sequence and secondary structure of MarA and the DNA oligomer used for crystallization. (A) Sequence of MarA aligned with SoxS, Rob, and AraC by using the pileup program (31). The secondary structural elements were assigned with the dssp program (32) and the HTH motifs represented by shaded boxes. Residues are enclosed in boxes according to their putative roles: white boxes for the hydrophobic core residues of the HTH motif and black-shaded boxes for the residues determining the sequence specificity. Asterisks mark MarA residues that interact with the phosphate backbone group of DNA. Residue numbers above the alignment correspond to the MarA sequence. (B) Double-stranded oligonucleotide used for crystallization. Sequences that are protected by DNase footprinting (5) are enclosed in a box. The underlined sequence is not a part of the mar promoter sequence but was required for crystal formation.