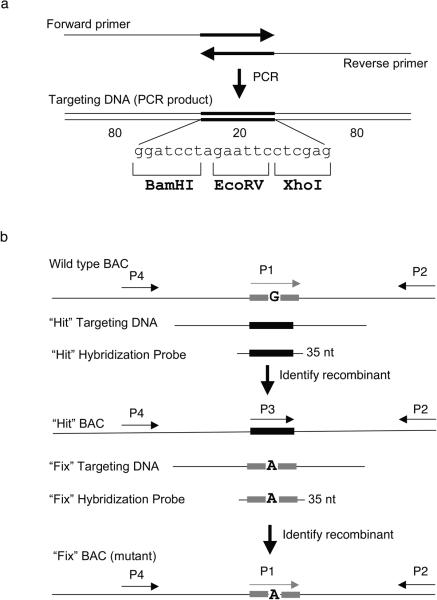
Figure 6. Two-step “hit & fix” method to generate subtle mutations using single stranded short PCR product or oligonucleotides as targeting vector.
(a) The single-stranded oligonucleotides containing 160 bases of homology and 20 unique bases are generated by using two 100-mer oligonucleotides in a PCR reaction. The two 100-mer oligonucleotides have 20 complementary bases (in this case the 20 bp contains restriction sites BamHI, EcoRV, XhoI) at the 3′ end. The 180 bp PCR product can be denatured to obtain single-stranded oligonucleotides that can be used as targeting construct. (b) Schematic representation of the two steps involved in “hit and fix” method to generate subtle alterations (e.g. G to A) without the use of a selectable marker. In step 1, a 180-mer single-strand oligonucleotide is used to replace 20 nucleotides (gray box) around the target site with 20 heterologous nucleotides (black box). Recombinants can be identified by colony hybridization using an end-labeled 35-mer oligonuclotide that can specifically anneal only to the recombinant DNA. A primer set specific for the heterologous “hit” sequence (P3 and P2) can be used to confirm the presence of recombinant clones by PCR. A second primer set (P1 and P2) can be used as a control to amplify only the non-recombinant DNA. Generation of a correct recombinant clone can be confirmed by digesting with BamHI, EcoRV or XhoI the PCR product (~300–500 bp) of primers P2 and P4. In step 2, the 20 nucleotides are restored to the original sequence, except for the desired mutation. Such clones can be identified by colony hybridization using a 35-mer oligonucleotide as probe and further confirmed by PCR amplification using primers P1 and P2, by testing for loss of the restriction sites inserted in step 1, by digesting the PCR product of primers P2 and P4, and by sequencing.