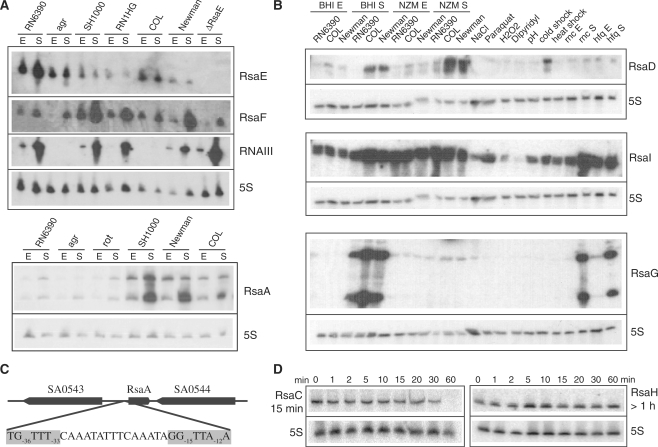
Figure 1.
The expression of Rsa RNAs as monitored under various stresses, culture conditions, and in various S. aureus strains. (A) Northern blot analysis of RsaE, RsaF and RsaA in different S. aureus strains: RN6390 (ΔrbsU, σB−), RN6911 (Δagr, σB−), SH1000, RN1HG, COL and Newman (σB+). The strains LUG1430 (ΔrnaE) and LUG1160 (Δrot) are isogenic to RN6390. Total RNAs were prepared from cells grown in BHI medium and stopped at the exponential (E) or stationary (S) phases of growth. Staphylococcus aureus RNAIII was used as positive control, and 5S rRNA was probed as a quality RNA control. (B) Northern blot analysis of RsaD, RsaI and RsaG in various S. aureus strains (RN6390, COL, Newman, LUG774 (RN6390-Δrnc) and LUG911 (RN6390-Δhfq)) grown in BHI or NZM at exponential (E) and stationary (S) phases of growth. The COL strain was grown under various stress conditions: osmotic stress (NaCl), oxidative stress (H2O2, paraquat), iron chelating agent (dipyridyl), acidic pH, cold shock (25°C) and heat shock (42°C). (C) Genomic organization of the rsaE gene with the σB-binding site; the consensus sequence is highlighted. (D) Northern blot analysis of the half-lives of RsaC and RsaH. The cells were treated with rifampicin and total RNAs were extracted after 1, 2, 5, 10, 20, 30 and 60 min at 37°C in BHI. 5S rRNA was probed to quantify the yield of RNA in each lane.