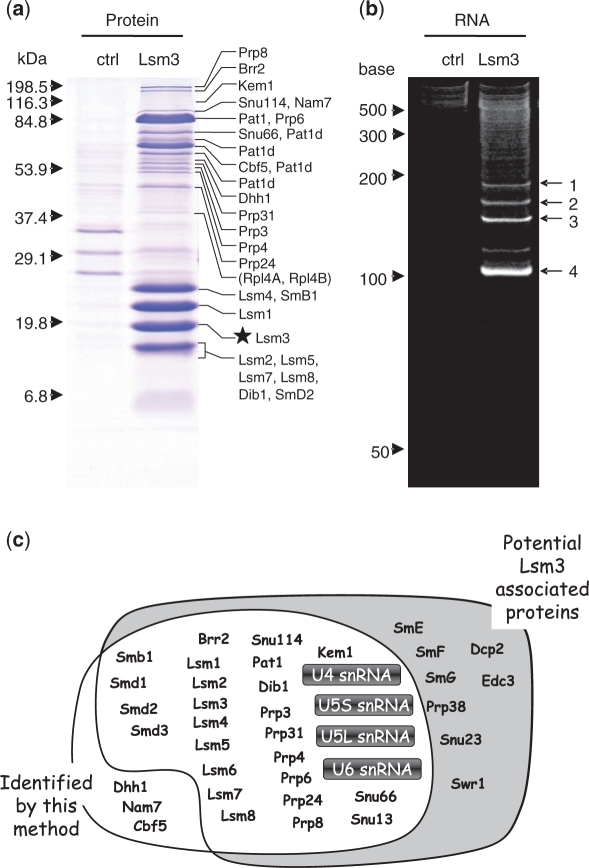
Figure 6.
Protein and RNA composition of the purified yeast Lsm3-associated snRNP complex. (a) SDS–PAGE profile of the control (ctrl) and the Lsm3-associated RNP complex (Lsm3) pulled down with TAP-tagged Lsm3 as affinity bait (visualized by Coomassie brilliant blue staining). The control experiment was performed using TAP-tagged Ist3, which forms a pre-spliceosomal RNP complex that shares no protein/RNA components with the Lsm3-associated complex (78). The molecular mass markers are indicated on the left and the proteins identified by LC-MS/MS analysis of individual bands excised from the gel are shown on the right. The star indicates the position of TAP-tagged Lsm3 protein. Note that the gel-based proteomic analysis identified 25 known protein cofactors of the yeast Lsm3 complex and several Pat1-related proteins with apparently different molecular size (designated as Pat1d; Supplementary Table S1), as well as other yeast proteins, Rpl4A and B, that were also detected in the control Ist3 complex and thereby considered as contaminants in our Lsm3 complex preparation (indicated in parentheses). (b) PAGE profile of RNA components in the control (ctrl) and the Lsm3-associated complex (Lsm3). The bands 1–4, visualized by SYBR Gold staining containing 4–10 ng RNA (100–300 fmol), were subjected to LC-MS analysis. The RNA size markers are indicated on the left. (c) A schematic representation of the yeast Lsm3-associated proteins and RNAs identified in this study (white background) and their overlap with the known components reported in previous studies (grey background). Note that the MS-based technology reported here identified most protein cofactors and four U snRNAs (highlighted) of this pre-spliceosomal RNP complex. Detailed data for the protein and RNA analyses are given in Supplementary Figure S2 and Supplementary Tables S1–6.