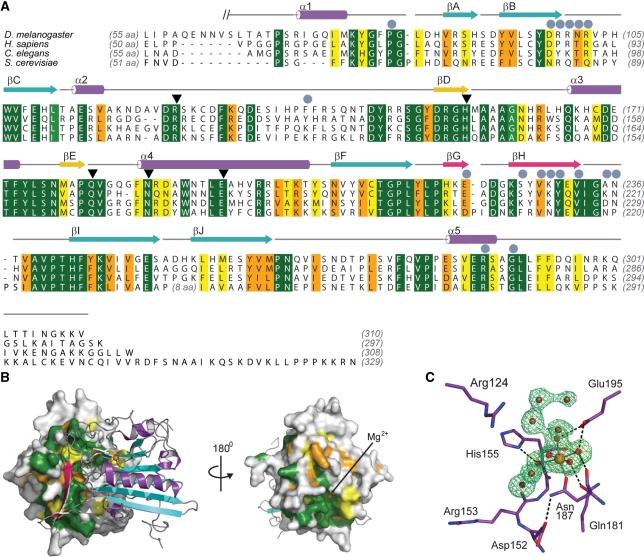
Figure 2.
Sequence conservation of EndoG. (A) Sequence alignment of EndoG homologues for D. melanogaster (NP_610737), Homo sapiens (NP_004426), Caenorhabditis elegans cps–6 (NP_491371) and S. cerevisiae Nuc1p (NP_012327). Identical residues are colored dark green and according to the decreasing similarity from light green through orange to yellow. Secondary structure elements are colored according to Figure 1. Helices are drawn as cylinders and strands as arrows. Residues which are involved in formation of the dEndoG homodimer interface are labeled with gray circles and residues directly involved in catalysis are marked with black triangles. (B) Conserved patches at the molecular surface of dEndoG. Left: A surface representation of one dEndoG monomer is colored according the amino acid sequence alignment in (A), and the second molecule is drawn as a ribbon model similarly to Figure 1. Right: the dEndoG homodimer turned by 180°, showing the conserved patch around the active site of dEndoG. (C) Active site of dEndoG. Residues involved in metal ion coordination and/or catalytic function are shown as stick models, the central Mg2+ ion as an orange sphere and water molecules from the metal ion hydration shell and solvent water molecules are shown as red spheres. An electron density map contoured at 3.0 σ for the omitted metal ion and water molecules is shown as a green mesh.