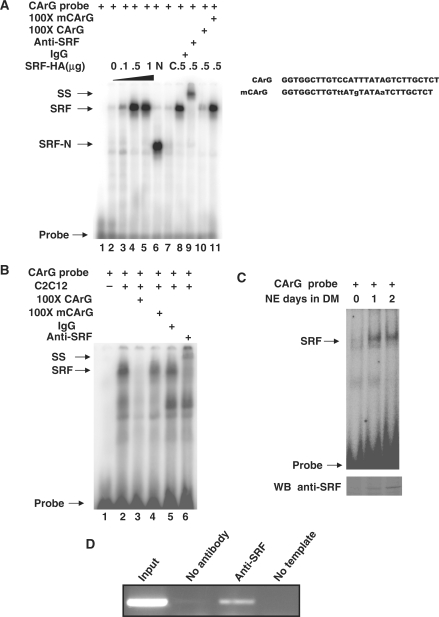
Figure 5.
Identification of SRF binding to the CArG site of SMYD1 promoter in vitro and in vivo. (A) Nuclear extracts from 293T cells transfected with SRF-HA (lanes 2–5 and 8–11), SRF-N (lane 6) or SRF-C (lane 7) constructs were incubated with [32P] radiolabeled CArG oligonucleotides and competed with 100× unlabeled cold or mutant competitor (lane 10 and 11, respectively), non-specific IgG (lane 8), anti-SRF antibody (lane 9) as described in ‘Materials and Methods’ section. Oligo sequence used for EMSA is listed on the right side. (B) Nuclear extracts from C2C12 myotubes were incubated with [32P] radiolabeled CArG oligonucleotides (lane 2) and competed with 100-fold unlabeled cold wild type or mutant competitor (lanes 3 and 4, respectively), non-specific IgG (lane 5), anti-SRF antibody (lane 6) as described in ‘Materials and Methods’ section. (C) Nuclear extracts from C2C12 cells in different time point of differentiation were incubated with [32P] radiolabeled CArG oligonucleotides as described in ‘Materials and Methods’ section. The protein level of SRF was demonstrated with Western blots with SRF specific antibody. (D) ChIP analysis of SRF with Smyd1 promoter region. C2C12 cells were differentiated for 2 days, harvested, and processed. After IP of the cross-linked complexes, the chromatin was analyzed by PCR. PCR amplified a 341-bp region containing the SRF-binding site, as described in ‘Materials and Methods’ section. Single PCR product was obtained and the identity of the band was confirmed by sequencing (data not shown). Results in these assays are from triplicate experiments. NE, nuclear extract; SS, super-shift; DM, differentiation medium; WB, western blot.