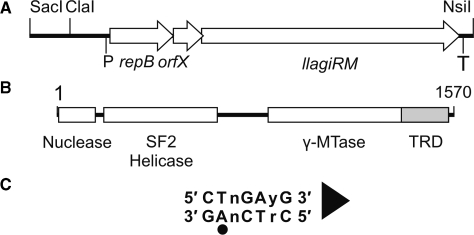
Figure 1.
The LlaGI RM enzyme. (A) The SacI-NsiI region of pEW104 identified previously as sufficient for in vivo RM activity (1). The approximate locations of the putative promoter (P) and terminator (T) of the operon are indicated. (B) The llagiRM gene product is a single polypeptide, multi-domain protein (1). Protein domain boundaries were estimated using secondary structure prediction and alignment with LlaBIII (not shown). Target Recognition Domain (TRD). (C) Recognition sequence of LlaGI. The adenine residue that is methylated is indicated by a circle. The arrowhead defines the directionality of the site according to cleavage (Figure 4) and translocation assays (10). The ‘top strand’ sequence (i.e. that starting with CT) is quoted throughout.