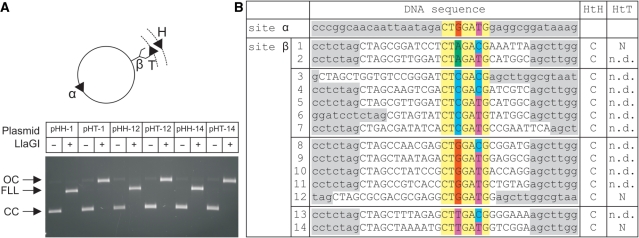
Figure 3.
Cleavage of LlaGI recognition sequences. (A) A library of two-site DNA substrates were generated from pOne (Figure 4) by cloning different LlaGI sequences (Site β – see B). Site α was constant in each case. For three of the sequences both directly-repeated and indirectly-repeated copies of Site β were tested. Example agarose gel shown where CC is Covalently closed Circular substrate DNA, OC is Open Circle nicked intermediate and FLL is the Full Length Linear product cleaved in both strands. DNA and LlaGI were incubated for 10 min. (B) Sequences of sites tested with outcome for indirectly-repeated (HtH) and directly-repeated (HtT) arrangements of sites: C, dsDNA cleavage; N, nicking only; n.d., not determined. pOne sequence is highlighted in grey, cloned sequence in white, constant LlaGI sequence in yellow and degenerate positions in red (dG), green (dA), blue (dC), purple (dT). Sequence shown is the ‘top strand’ as defined in Figure 1C.