Figure 1.
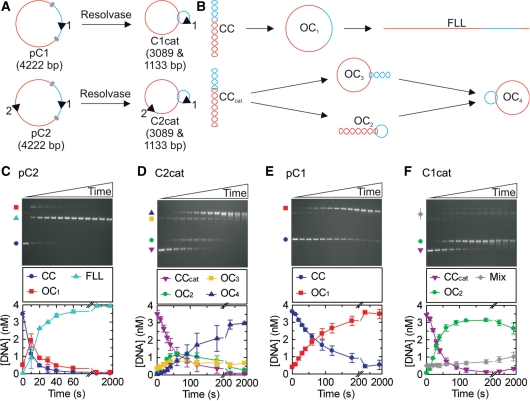
LlaGI endonuclease activity measured on DNA catenanes. (A) The two plasmid substrates used to form catenanes in the presence of Tn21 resolvase. LlaGI recognition sequences are represented as arrowheads where the orientation is 5′-CTnGAyG-3′. Res sites for Tn21 resolvase are shown as grey blocks whilst the different DNA domains that are separated by recombination are shown in red and blue. (B) Possible DNA species formed when LlaGI is incubated with the plasmid or catenane substrates. Each arrow represents the cleavage of one DNA strand. CC, covalently closed circular DNA (negatively supercoiled); OC1, open circle DNA (‘nicked’); FLL, full-length linear DNA; CCcat, covalently closed circular catenane DNA. For the catenanes, OC2 is nicked in the small ring only, OC3 is nicked in the large ring only and OC4 is nicked in both rings. Example gels and average cleavage profiles are shown for: (C) two-site plasmid DNA pC2; (D) two-site catenane DNA C2cat; (E) one-site plasmid DNA pC1; and (F) one-site catenane DNA C1cat. Reactions contained 200 nM LlaGI, 4 nM DNA and 4 mM ATP (‘Materials and Methods’ section). ‘Mix’ in (F) represents the combination of OC3, OC4 and dimeric DNA products. Time points were collected at 0, 10, 20, 30, 40, 60, 90, 120, 180, 300, 600, 1200 and 1800 s.