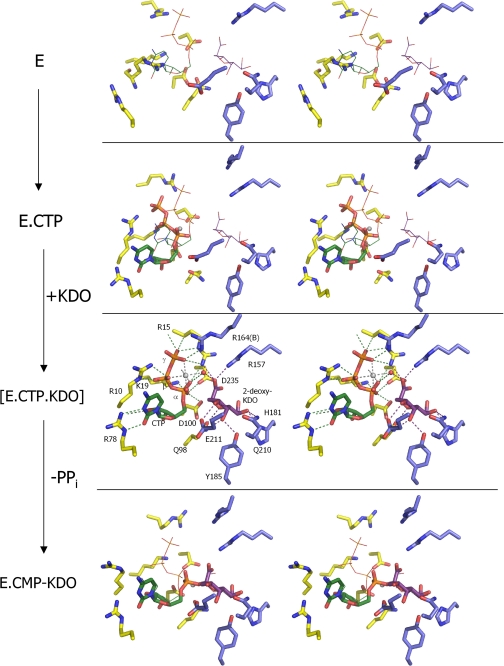
FIGURE 5.
Crystallographic snapshots along the KdsB reaction path. Key residues from the active site of four different E. coli CMP-Kdo synthetase crystal structures are displayed with residues derived from the dimerization domain in blue carbons and those derived from the CTP-binding domain in yellow carbons. Mg2+ ions (when present) are shown as gray spheres. The positions of the bound ligands from the CTP-2β-deoxy-Kdo-KdsB crystal structure are shown as thin lines in the other crystal structures for easy comparison. The ligand-free enzyme structure represents E (this work). although the CTP-bound E. coli capsule-specific CMP-Kdo synthetase structure (PDB code 1GQ9, see Ref. 14) represents the CTP-E complex. The ternary enzyme complex E-CTP-Kdo is mimicked by the CTP-2β-deoxy-Kdo-KdsB crystal structure (this work), and the E-CMP-Kdo product-bound enzyme structure is represented by the CMP-Kdo-bound E. coli capsule-specific CMP-Kdo synthetase structure (PDB code 1GQC, see Ref. 14).