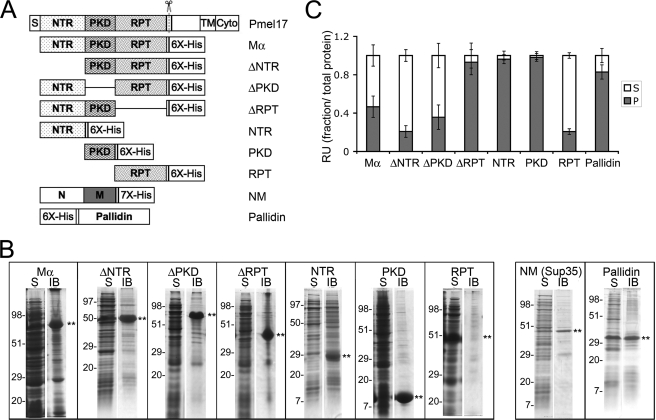
FIGURE 3.
Solubility of recombinant Pmel17 His-tagged constructs. A, schematic diagram of full-length Pmel17 and C-terminally His-tagged recombinant lumenal domain fragments. Δ denotes that the indicated domain has been deleted from full-length Mα. His6, hexahistidine tag on recombinant proteins. Also shown is the His7-tagged prion-forming subdomain (NM) of the yeast prion protein Sup35, used as a positive amyloid control, and the His6-tagged full-length Pallidin, used as a negative control. B, partitioning into soluble and insoluble bacterial fractions. BL21 E. coli expressing the different proteins indicated in A were harvested and processed as described under “Experimental Procedures.” The soluble (S) and insoluble IB fractions were separated by SDS-PAGE and analyzed by Coomassie Blue staining. Asterisks denote the position of the induced protein, and the migration of molecular weight standards is indicated to the left of each pair of lanes. Note that all of the Pmel17-derived proteins are found predominantly in the IB fraction with the exception of the RPT, which is found predominantly in the soluble fraction. C, sedimentation. Each of the recombinant proteins was solubilized from inclusion bodies in guanidine HCl and affinity-purified by His-bind chromatography. Affinity-purified protein was diluted out of the denaturant into physiological buffer and allowed to refold overnight with agitation at 37 °C. Aliquots were fractionated into a soluble supernatant (S) and insoluble pellet (P) by centrifugation at 100,000 × g for 1 h at 4 °C. Total, supernatant, and pellet fractions were analyzed by SDS-PAGE, stained with Coomassie Blue, and image scanned; the relative amount of protein in each fraction, assessed as signal intensity, was determined using ImageQuant software. The mean fraction of protein in the supernatant and pellet fractions relative to the total is plotted ± S.D. n = 3 for Pallidin, n ≥ 4 all others. RU, relative unit.