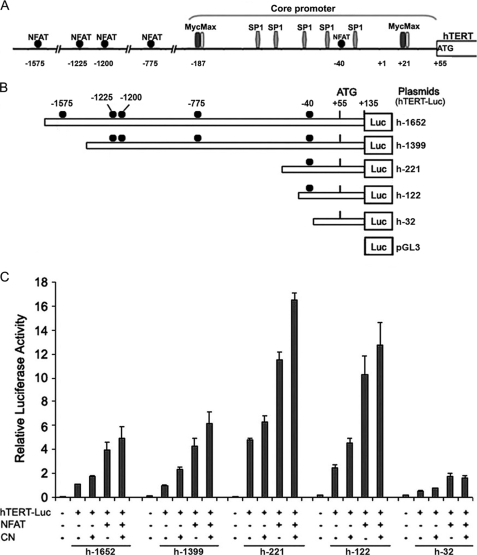
FIGURE 3.
Schematic representation of reporter plasmids and transcriptional activity of the hTERT promoter. A, schematic representation of the full-length hTERT promoter sequence used in this study. Potential NFAT-binding sites are indicated, as are E-boxes and SP1-binding sites in the promoter core. B, the hTERT-Luc reporter plasmids were generated by inserting the 1787-bp DNA and 5′-truncated fragments of the hTERT promoter upstream of the +1 transcription initiation site into the luciferase (Luc) reporter vector pGL3-basic in the sense orientation. The locations of NFAT-binding sites are indicated as the number of their first base upstream of the +1 transcription initiation site. The name of each reporter construct was assigned according to the nucleotide number at the 5′-end of the inserted promoter sequences. C, telomerase-negative GM847 cells were cotransfected with 1.5 μg of the different hTERT-Luc reporter plasmids in the absence (−) (3 μg of pEFTAG) or presence (+) of the HA-NFAT1 expression vectors (3 μg of pEFTAG-mNFAT1c), and in the absence (−) (0.6 μg of SRα) or presence (+) of the constitutively active calcineurin (CN) expression vectors (0.6 μg of ΔCAM-AI). For each transfection, luciferase activity was normalized to the Renilla luciferase activity due to cotransfected pRL-TK (0.25 μg). The relative activity of each construct is expressed as the ratio of its activity to that of h-1652 cotransfected with the empty versions of the NFAT and CN expression vectors (−). The negative control in these experiments involved cotransfection of the pGL3-basic vector with the empty expression vectors (−). The mean and standard deviation of at least three experiments are shown for each construct.