Abstract
Aims
Cardiac hypertrophy is a risk factor independent of blood pressure; however, the mechanisms that distinguish pathological remodelling due to local cues from pressure overload are unresolved. This study was aimed at discovering a novel gene expression mechanism in heart failure.
Methods and results
In angiotensin II type 1 receptor (AT1R) transgenic mice (TG), we found a significant increase of mRNA and total STAT3 (T-STAT3) protein, but not STAT3 phosphorylated at residues Y705 and S727. A net increase in nuclear accumulation of this unphosphorylated form of STAT3 (U-STAT3) correlated with the development of cardiac hypertrophy and dysfunction, which are associated with abnormal expression of osteopontin and regulator of G protein signalling 2 genes. Nuclear accumulation of U-STAT3 is induced by angiotensin II treatment in neonatal cardiac myocytes, fibroblasts, and AT1R-expressing human embryonic kidney 293 (HEK-AT1R) cells. Chromatin immunoprecipitation demonstrated that U-STAT3 binds to the target gene promoter, and siRNA-mediated knockdown of STAT3 expression significantly altered the expression of target genes in HEK-AT1R cells. T-STAT3 in TG mouse hearts and the phosphorylation-deficient Y705F mutant STAT3 in HEK-AT1R cells physically interacted with transcription co-activator p300.
Conclusion
Chronic activation of AT1R induces unregulated expression of the Stat3 gene, leading to nuclear accumulation of U-STAT3, which significantly correlated with progression of cardiac hypertrophy.
Keywords: Hypertrophy, Angiotensin II type 1 receptor, Unphosphorylated STAT3, Heart failure
1. Introduction
Despite significant therapeutic advances, morbidity and mortality in heart failure (HF) remain unacceptably high. The single most powerful predictor for the development of HF is the presence of left ventricular hypertrophy (LVH).1 Signal transduction induced by local neuronal and endocrine pathways in myocytes can cause reactivation of genes whose expression is normally restricted or strictly regulated, leading to cardiac hypertrophy and HF.2 Thus targeting signal transduction pathways to reverse or prevent cardiac hypertrophy is a goal of paramount importance in reducing HF.
Angiotensin II (Ang II) and its type-1 receptor (AT1R) are key components of the renin–angiotensin system (RAS) that regulate blood pressure, modulate salt/water balance, and promote growth and proliferation of cells. Abnormal regulation of RAS is linked to hypertension, cardiac hypertrophy, and HF.3,4 Apart from its systemic effects, local growth promoting activity of the cardiac RAS is revealed, with the evidence that selective blockade reduce LVH.3,4 Mechanical stretch can locally activate myocyte AT1R signalling and can therefore, directly induce cardiac hypertrophy.5 Increase of AT1R has been reported in the heart after myocardial infarction.6 A number of transgenic animal models designed to overexpress AT1Rs in cardiac myocytes demonstrated the predicted influence of local RAS, but the signal transduction mechanisms are not clearly established (see Supplementary material online, Table S1).
In addition to the G protein, Gq, AT1R stimulates several tyrosine kinases including JAK, Src, and EGFR.7 Activation of the JAK/STAT pathway by Ang II has been observed in cardiac myocytes and fibroblasts.8 Ang II effects in myocardial infarction result at least in part from AT1R-mediated JAK/STAT signalling,9 and adverse cardiomyocyte remodelling due to STAT3-dependent activation of local RAS.8 Latent STATs reside in the cytoplasm until activated via tyrosine phosphorylation (Y705 in STAT3), which leads to their dimerization and nuclear translocation.10 Serine phosphorylation (S727 in STAT3) is needed for maximal activation of transcription, but not for DNA binding.11,12 All seven STAT family homologues are expressed in the heart, but their functions are not clearly defined. Therefore, intricacies of regulation of STAT3 in cardiac hypertrophy and HF require further study.
Unlike phosphorylated STATs, the unphosphorylated STATs (U-STAT) drive pathological gene expression in chronic diseases.13,14 Whether U-STAT signalling is involved in cardiac hypertrophy is unknown. We show that expression of the Stat3 gene is induced in AT1R transgenic mice (TG), which promotes persistent nuclear accumulation of STAT3 not phosphorylated at residues Y705 and S727 (referred to as U-STAT3), significantly correlated with cardiac hypertrophy and associated pathogenic gene expression. Our finding emphasizes a novel concept that U-STAT3 acts as a ‘rogue’ transcription factor and drives AT1R-mediated cardiac hypertrophy and may affect transition to HF.
2. Methods
2.1. Mouse model of cardiac restricted expression of hAT1R
The αMHC-AT1R TG were kindly provided by Paradis et al.15 in C57BL/6 genetic background. The relative level of AT1R in the cardiac membrane was estimated to be ∼100-fold higher in TG relative to non-transgenic (NTG) mice.16 The animal use was according to the Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (NIH Publication No. 85-23, revised 1996). All experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee.
2.2. Sample harvest and histological analysis
Age- and sex-matched TG and NTG littermates were randomly selected and an echocardiogram was performed in a blind fashion. The hearts were harvested and histological analysis was performed as described previously.17
2.3. Primary culture of neonatal cardiac myocytes and fibroblasts
Mouse neonatal cardiac myocytes and fibroblasts were cultured from 1- to 2-day-old NTG mice as described previously.18 The cardiomyocytes were seeded into poly-d-lysine coated chamber slides and immunofluorescence staining was performed after optimal treatments. Third passage cardiac fibroblasts were used for all experiments.
2.4. Microarray analysis of gene expression
Total RNA was extracted from mouse hearts with RNeasy fibrous tissue kit (Qiagen) or from cultured cells using RNeasy Mini kit (Qiagen). Approximately 15 µg of RNA was used to obtain cRNA according to standard Affymetrix protocols. The cRNA was hybridized with human (U133A) or mouse (U74Av2) arrays and data were analysed using Affymetrix gene chip software. The expression of genes in TG mice was compared with NTG as the baseline. In AT1R-overexpressing human embryonic kidney 293 cells (HEK-AT1R), after stimulation with Ang II, gene expression was compared with untreated cells as the baseline.
2.5. Western blot, immunoprecipitation, and real-time RT–PCR
Preparations of protein and RNA extracts were performed per the protocols provided by the manufacturer. Western blot, immunoprecipitation, and real-time RT–PCR were performed as described previously.17
2.6. Plasmid vector construction, gene transfection, and gene knockdown
HA-tagged human AT1R was cloned into pcDNA3 vector; Flag-tagged mouse STAT3 (Flag-WT-STAT3) and Y705F mutant STAT3 (Flag-Y705F-STAT3) were cloned into pcDNA6. Pre-designed siRNA targeting human STAT3 (validated, ID# 6793, Ambion) and control siRNA were transfected into HEK-AT1R cells using Silencer siRNA Transfection II Kit (Ambion).
2.7. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using the EZ-Chip™ kit (Millipore). STAT3-associated chromatin fragments were immunoprecipitated using anti-FLAG, anti-pY705-STAT3, and anti-pS727-STAT3 antibodies. Eleven pairs of primers were designed (see Supplementary material online, Table S4) based on the 8000 bp upstream sequence of human OPN (AF052124) for amplification of immunoprecipitated promoter fragments.
2.8. Statistical analysis
All data are expressed as mean ± SEM. Statistical analyses between groups were performed with one-way ANOVA followed by unpaired Student t-test. A P-value less than 0.05 was considered statistically significant.
3. Results
3.1. Anomalous STAT3 expression associated with AT1R-induced cardiac hypertrophy and dysfunction
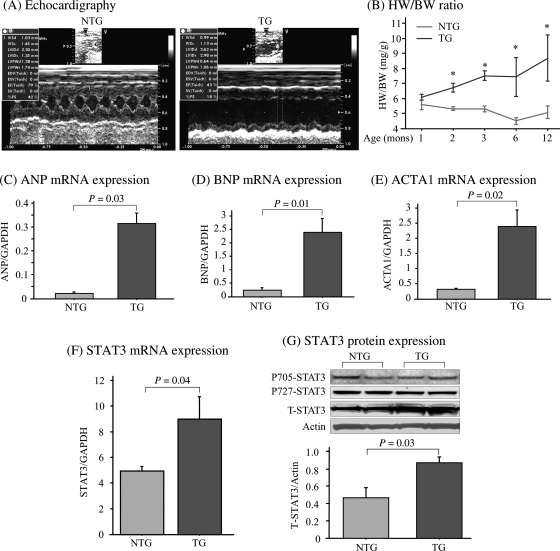
We observed LV enlargement and a decrease of myocardial function in the TG mice (Figure 1A and see Supplementary material online, Table S2) in agreement with previous finding.15 Average size of the myocytes increased significantly (P = 0.0004) in the TG mice, suggesting cardiomyocyte hypertrophy (see Supplementary material online, Figures S1 and S2). Post-mortem pathology demonstrated that both ventricles and atria were enlarged in the TG hearts, and the ratio of heart weight to body weight (HW/BW) was significantly increased in TG mice (see Supplementary material online, Table S2). To further confirm this finding, age-dynamic change in cardiac growth and function was examined at 1, 2, 3, 6, and 12 months of age in both groups. A significantly higher HW/BW was found in TG mice at 2 months and throughout the monitoring period, but not in NTG mice (Figure 1B). Thus, development of cardiac hypertrophy and dysfunction in TG mice is due to hAT1R transgene expression and not an age-related effect. The mRNA levels of representative foetal genes, including atrial natriuretic peptide, brain natriuretic peptide, and alpha 1 skeletal muscle actin (ACTA1), were significantly increased in TG mice (Figure 1C–E), providing further evidence for AT1R-induced hypertrophy.
Figure 1.
Anomalous STAT3 expression associated with AT1R-induced cardiac hypertrophy and dysfunction. (A) Representative echocardiogram images of 3-month-old TG and NTG mice. (B) Dynamic changes of HW/BW ratio with age. *P < 0.01 TG vs. NTG (n = 5 for each age in each group). Foetal gene expression including atrial natriuretic peptide (ANP) (C), brain natriuretic peptide (BNP) (D), and ACTA1 (E) was analysed by real-time RT–PCR, and data were expressed as ratio to GAPDH expression (n = 4). (F) Real-time PCR, and (G) immunoblotting assay of STAT3 expression in the heart tissue (3 months old). Actin was used as a loading control and bar graph shows ratio of T-STAT3 to actin. pY705-phosphorylated tyrosine 705; pS727-phosphorylated serine 727; T-total.
To understand the AT1R signalling mechanisms mediating cardiac hypertrophy, we examined predicted pathways, for example, MAPK signalling cascades—ERK1/2, p38, and JNK signalling. No differences were observed between TG and NTG mice (data not shown). However, the tyrosine kinase-activated transcription factor, STAT3, was markedly increased in TG mice, at both the mRNA and protein levels (Figure 1F and G). Surprisingly, the total STAT3 (T-STAT3) protein level was increased, but STAT3 phosphorylated at tyrosine 705 and serine 727 did not, suggesting a net increase of U-STAT3 in the TG mice. The increased STAT3 mRNA in TG mice is correlated with the increased U-STAT3, suggesting that the increase of U-STAT3 results from transcription.
3.2. Nuclear accumulation of U-STAT3 in TG mouse heart cells and correlation with cardiac hypertrophy
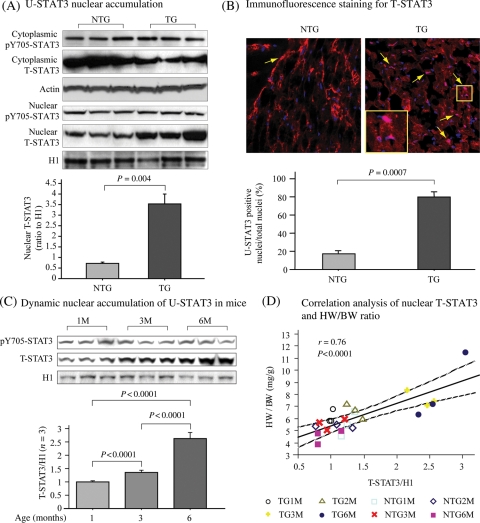
Nuclear translocation initiated by Tyr705 phosphorylation (pY705-STAT3) is central to the transcription factor function of STAT3. By analysis of nuclear and cytosolic fractions, we found a significant increase of nuclear T-STAT3 in TG mice (Figure 2A). The level of pY705-STAT3 was not increased in the nuclear fraction, suggesting a net increase of U-STAT3 in the nuclei of TG mouse hearts. Immunofluorescence (Figure 2B) and immunohistochemical (see Supplementary material online, Figure S2) analyses demonstrate T-STAT3 accumulation in cardiomyocyte nuclei in TG mice. Additionally, T-STAT3 stained nuclei in the TG hearts was about four-fold higher than NTG. The negative correlation determined between nuclear accumulation of T-STAT3 and fractional shortening (P < 0.0001, r = −0.985, data not shown) was significant, suggesting that nuclear U-STAT3 may play an important role in AT1R-induced cardiac dysfunction.
Figure 2.
Nuclear accumulation of U-STAT3 in TG mouse cardiac cells is associated with the development of cardiac hypertrophy. (A) Immunoblot analysis of STAT3 and pY705-STAT3 in the nuclear and cytoplasmic fractions from heart tissue. (B) Representative immunofluorescence staining of a heart tissue section, red indicated staining of T-STAT3, nuclei were stained with DAPI. Bar graph shows the ratio of T-STAT3 positive nuclei to the total nuclei in the same area. Arrows indicate representative T-STAT3 positive nuclei. The inset shows merge of red and blue yielding pink colour. (C) Age-dynamic change of nuclear STAT3 by immunoblotting assay. Histone H1 was re-blotted as a loading control. Bar graph shows statistical analysis of ratio of nuclear T-STAT3 to histone H1 (n = 3). (D) Correlation analysis between HW/BW and nuclear T-STAT3 expression. pY705-phosphorylated tyrosine 705; T-total; HW: heart weight; BW: body weight; M: months.
Age-dependent nuclear accumulation of U-STAT3 was significantly increased in TG mice (Figure 2C). At 1 month, the pY705-STAT3 and T-STAT3 levels were similar in TG and NTG mice. Their levels did not change significantly in the NTG mice up to 6 months (data not shown). However, T-STAT3 but not pY705-STAT3, significantly increased at 3 and 6 months compared with 1 month, and at 6 months compared with 3 months in the TG, indicating age-dynamic nuclear accumulation of U-STAT3 in the TG mice. The positive correlation between the nuclear accumulation of T-STAT3 and HW/BW (Figure 2D) determined was significant, suggesting that the nuclear accumulation of U-STAT3 plays a role in the progression of cardiac hypertrophy.
3.3. Nuclear accumulation of U-STAT3 upon chronic activation of AT1R in cardiac cells and in HEK-AT1R cells
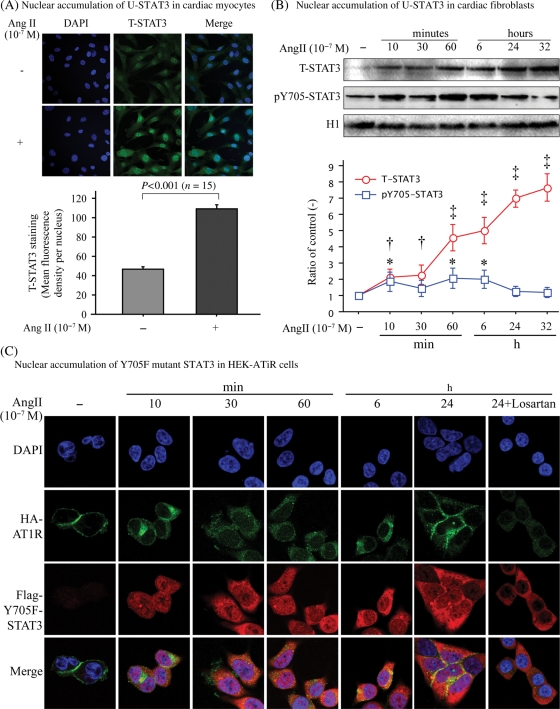
To evaluate the conditions that cause endogenous AT1R to initiate nuclear accumulation of U-STAT3, different types of cells were stimulated with Ang II for different times and nuclear accumulation of U-STAT3 was compared. Both pY705-STAT3 and T-STAT3 were found in the nuclear fraction after Ang II stimulation of neonatal cardiomyocytes and fibroblasts (Figure 3A and B). The pY705-STAT3 showed two peaks (10 and 60 min) then returned to baseline at 32 h; however, the T-STAT3 continually increased ≈seven-fold over baseline at 32 h. The AT1R antagonist, losartan, blocked this Ang II effect (data not shown). We conclude that chronic activation of AT1R causes persistent nuclear accumulation of T-STAT3.
Figure 3.
Kinetics of nuclear accumulation of U-STAT3 in cardiac cells and in HEK-AT1R cells. Cardiac neonatal myocytes (A), fibroblasts (B), and Flag-Y705F-STAT3/HA-AT1R double transfected HEK293 cells (C) were stimulated with Ang II for different times as indicated. (A) Cardiomyocytes were stimulated with Ang II for 24 h and stained for T-STAT3. Green indicates positive staining. Quantification analysis of nuclear fluorescence density was performed using NIH Image. (B) Nuclear fractions were immunoblotted for pY705-STAT3, T-STAT3, and histone H1. Bands were analysed and fold changes of the density to non-treated control with time were shown in the line graph. *P < 0.05, †P < 0.01. ‡P < 0.001. (C) Immunocytofluorescence staining of Flag-Y705F-STAT3 (red) and HA-AT1R (green) double transfected HEK293 cells, which were treated with Ang II with/without losartan for different times. Nuclei were stained with DAPI.
The non-cardiac human cell line, HEK-AT1R, reproduced the same phenomenon, hence was used for the mechanistic studies described below. We used the Flag-Y705F-STAT3 as the U-STAT3-surrogate and constructed HEK lines stably expressing Flag-Y705F-STAT3 and HA-AT1R. Ang II stimulated dynamic trafficking of AT1R and nuclear accumulation of Flag-Y705F-STAT3, providing direct evidence that without Y705-phosphorylation, STAT3 accumulates in the nucleus (Figure 3C). The Flag-Y705F-STAT3 mutant was not Ser727 phosphorylated in HEK cells (data not shown). Nuclear accumulation of U-STAT3 surrogate disappeared with losartan treatment. The cells stably expressing Flag-WT-STAT3 and HA-AT1R also showed similar results (data not shown).
3.4. Ang II/AT1R-induced expression of candidate U-STAT3 regulated genes
Previously, Yang et al.14 reported U-STAT3-driven genes in mouse and human cells. We used their dataset to ‘filter out’ genes for evaluating their potential regulation by U-STAT3 in our experimental models.
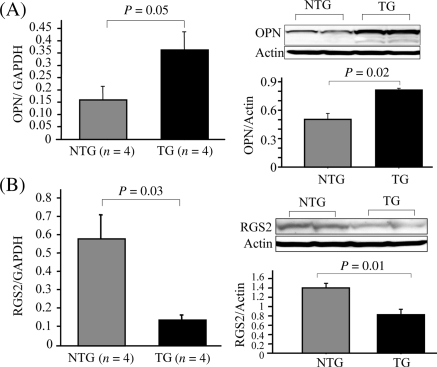
Expression of 775 genes was altered >2-fold (data not shown) in HEK-AT1R cells stimulated with Ang II for 24 h compared with un-stimulated cells. Twenty-four genes in this dataset matched the U-STAT3 driven genes identified in human mammary epithelial cells (see Supplementary material online, Table S3), suggesting that they may be regulated by U-STAT3 in human cells. A similar comparison between differentially expressed genes in the TG mouse hearts and Stat3-null mouse embryonic fibroblasts overexpressing Y705F-STAT3 yielded potential mouse U-STAT3-target genes (see Supplementary material online, Table S3). Functional annotation of both sets of genes in the NCBI genome browser identified six genes potentially involved in HF pathogenesis. We examined the expression of two of six genes in mouse heart. Both mRNA and protein levels were significantly increased in TG mice for osteopontin (OPN), which is a well-known marker for severity of HF (Figure 4A).19 The mRNA and protein levels decreased in TG mice for regulator of G protein signalling 2 (RGS2), which has been reported to play an important role in negative regulation of G-protein-dependent cardiac hypertrophic signalling (Figure 4B).20 These data confirmed the validity of our prediction. We propose that differential expression of predicted U-STAT3 target genes correlate with cardiac hypertrophy and dysfunction in the hAT1R TG mice.
Figure 4.
Expression of U-STAT3 target genes in AT1R transgenic mice. Real-time RT–PCR and western blot analyses were performed to evaluate expression of the OPN (A) and RGS2 (B) in mouse hearts.
3.5. Regulation of target gene expression by U-STAT3
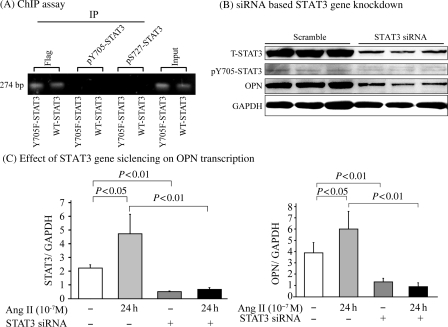
To demonstrate that U-STAT3 binds to the promoter to regulate its target genes, such as OPN, we performed ChIP assay. The primer set (forward) 5′-GGGCAAACTGATTCTGGATGACTC-3′ and (reverse) CCAGAGTAGGGGATTGAGGAGAAAC, successfully amplified the region from −1883 to −1610 of the human OPN gene promoter. The 274 bp PCR product was obtained when immunoprecipitation was performed using anti-Flag antibody. There were no PCR products detectable when the anti-pY705-STAT3 or anti-pS727-STAT3 antibodies were used, suggesting that the U-STAT3 binding site is located within the −1883 to −1610 region of the OPN promoter (Figure 5A). There were no PCR products when the isotype control antibodies were used for immunoprecipitation (data not shown).
Figure 5.
U-STAT3 regulates OPN expression. (A) Y705F-STAT3: Flag-Y705F-STAT3 and AT1R double transfected HEK293 cells, and WT-STAT3: Flag-wild-type-STAT3 and AT1R double transfected HEK293 cells were used for the ChIP assay employing specific antibodies as indicated. (B) HEK-AT1R cells were transfected with siRNA targeting human STAT3 or scrambled siRNA, and expression of STAT3 and OPN were analysed by immunoblotting assay. GAPDH was blotted as a loading control. Cells transfected with STAT3 siRNA or scrambled siRNA were treated with (+) or without (−) Ang II for 24 h, and STAT3 (C, left) and OPN (C, right) mRNA expression was assessed by real-time RT–PCR assay (n = 4).
To further demonstrate STAT3 regulation of OPN and RGS2 gene expression, endogenous STAT3 expression was knocked down in the HEK-AT1R cells using an established siRNA.21 The T-STAT3 was knocked down by ∼70% (maximum at 50 µM of siRNA) compared with scrambled RNA-treated cells. Effect on pY705-STAT3 was not as dramatic. Following STAT3 knockdown, a marked decrease of OPN (Figure 5B) and RGS2 (see Supplementary material online, Figure S3A) was found. Taken together, these experiments suggest that STAT3 binding to OPN promoter (RGS2 as well) is essential for expression.
STAT3 dependence of OPN and RGS2 regulation by Ang II was assessed by real-time RT–PCR assay. Ang II induced a significant increase in STAT3 mRNA in control and >70% knockdown was achieved in STAT3 siRNA treated samples. STAT3 knockdown diminished Ang II-induced OPN (Figure 5C) and RGS2 (see Supplementary material online, Figure S3B) expression, indicating that the regulation of OPN and RGS2 by AT1R is critically dependent on the availability of higher levels of STAT3.
3.6. P300/CBP associated with U-STAT3
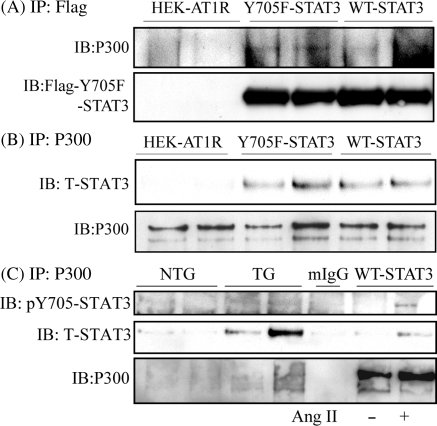
Association with CBP/p300 is an indicator of enhanced gene transcriptional activity of STATs.22 We examined potential association of p300 with U-STAT3 in our model. By immunoprecipitation of Flag-Y705F-STAT3 (U-STAT3) and Flag-WT-STAT3 (Figure 6A), or p300 (Figure 6B), we detected a complex formed between U-STAT3 and p300 in the HEK cells double transfected with AT1R and Y705F-STAT3. Similar experiments were performed in mouse heart extracts, in which a stronger interaction was found between T-STAT3 and p300 than between pY705-STAT3 and p300, in TG than in NTG (Figure 6C). Taken together, the interaction with p300 in vivo suggests involvement of U-STAT3 in gene transcriptional activity.
Figure 6.
Association of p300 and U-STAT3. Lysates from HEK-AT1R, Flag-Y705F-STAT3, and Flag-WT-STAT3 cells (A–C) or heart tissue (C) were used for co-immunoprecipitation assay as indicated. (C) Cell lysates from WT-STAT3 stimulated with (+) or without (−) Ang II were used as positive control; mIgG: mouse normal IgG, was used as a negative control. Y705F-STAT3: Flag-Y705F-STAT3 and HA-AT1R double transfected HEK293 cells; WT-STAT3: Flag-WT-STAT3 and HA-AT1R double transfected HEK293 cells.
4. Discussion
Hypertrophic signalling by cardiomyocyte-AT1Rs, in response to local RAS activation or myocyte-stretch, is a clinically relevant mechanism of cardiac hypertrophy.23 Mouse models overexpressing AT1R in cardiomyocytes display myocardial hypertrophy independent of BP and progressive pathological remodeling.15 However, molecular mediators of pathogenesis remain unknown. We discovered that nuclear accumulation of U-STAT3 was correlated with AT1R-induced cardiac hypertrophy and dysfunction (Figures 1–3) in the C57BL/6 mouse strain developed by Parades et al.15 The evidences obtained, U-STAT3 isolated from the nuclei of both TG mouse hearts and HEK-AT1R cells, was associated with transcription co-activator p300/CBP; U-STAT3 directly binds to a specific site on human OPN promoter and knockdown of U-STAT3 modulated expression of OPN, demonstrated that nuclear U-STAT3 promoted transcription (Figures 5 and 6). Potential target genes of U-STAT3 (see Supplementary material online, Table S3) include ACTA1, OPN, connective tissue growth factor (CTGF), and RGS2, which may influence pathogenesis of HF (Figures 4 and 5). This novel transcriptional mechanism involving p300/CBP-bound U-STAT3 has not been previously implicated in AT1R-induced HF pathogenesis.
Variation in contraction and remodelling observed in different AT1R transgenic models has lead to the speculation that the phenotype could be caused by embryonic myocardial defects or transgene overexpression and may not reflect remodelling in human HF.24 Therefore, we examined the ability of Ang II to mobilize U-STAT3 in normal cardiac myocytes and fibroblasts as well as the HEK-AT1R cells. Primary cardiac cells with endogenous levels and the HEK-AT1R with overexpression of AT1R, exhibited U-STAT3 accumulation upon chronic Ang II activation, similar in magnitude to that observed in the AT1R TG mice. Specificity was shown by treatment with the AT1R-selective antagonist, losartan. Expression of Y705F mutant STAT3 in the HEK-AT1R cells established that Tyr-705 phosphorylation deficient STAT3 is capable of translocation to the nucleus. Transfected wild-type and the mutant STAT3 were compared and the probable dominant-negative effect of the mutant was ruled out. These controls validate that the nuclear functions of U-STAT3 are not due to developmental artefacts, hAT1R over expression, or changes during progression of cardiac hypertrophy, and not an artefact induced by the transgene insertion.
There is considerable disagreement with regards to the role of STAT3 in the heart. Cardiac overexpression of STAT3 was reported to induce hypertrophy,25 which resembles low-dose Ang II-induced concentric hypertrophy26 and differs from our TG results. A constitutive increase of phosphorylated-STAT3 levels reported in the STAT3-model is consistent with a protective role of phosphorylated-STAT3 in adult hearts.27 In contrast, JAK/STAT signalling was associated with cardiac dysfunction during ischaemia and reperfusion.28 The STAT3 pathway plays a role in the hypertrophy of atria and fibrogenesis.8 In our study, the cardiomyocyte pathology was found in both ventricles and atria, where pathological remodelling occurred as reported earlier.15
Currently, mechanisms leading to U-STAT3 hyperactivation by Ang II/AT1R are incompletely understood. Chronic activation of AT1R coupled to JAK2-dependent or -independent pathways could phosphorylate STAT3, causing continual Stat3 gene transcription and resulting in a high intracellular concentration of U-STAT3.29,30 Nuclear translocation of STAT3, independent of tyrosine phosphorylation, requires interaction with importin-α,31 which is regulated by Ran GTPase.32 In hypertrophic myocardium, RGS2 may affect nuclear shuttling of U-STAT3 through its GTPase-activating function.33 We also suspect that the normal mechanisms responsible for clearance of U-STAT3 saturate and fail in HF, allowing U-STAT3 to accumulate in the nucleus.
Expression of certain genes in STAT3-deficient cells is driven by reconstitution with Y705F mutant after interleukin 1 and 6 stimulation.34 U-STATs can either bind to genomic DNA at a consensus GAS element (TTCNNNG/TAA) as a dimer or monomer, or at a half palindrome element (TTCNNNTAT) as a monomer.13 Moreover, U-STATs have been shown to partner with NF-κB p65 and IRF1 in binding to different hybrid DNA elements.13,34,35 Hence, U-STAT3 may partner with different factors depending on the cell type and sequence context, thereby regulating constitutive gene expression both positively and negatively. This may explain the differential effect of STAT3 on RGS2 expression in mouse hearts and HEK293 cells. U-STAT3 may activate genes that are normally not STAT3 regulated35 and may transform stringently regulated genes to express constitutively. As a result, more genes may be expressed and the expression of genes dysregulated as seen in HF. The function of U-STAT3 is clearly distinct from the absolute requirement for Tyr705 phosphorylation that enables dimerization and binding to GAS sequences to induce gene expression. The duration of phosphorylated STAT3 activity and the amount of U-STAT3 in cytosol ready to be activated in normal cells are critical determinants for normal functions of STAT3.
STAT3 has a large NH2-terminal coiled coil domain that allows the binding of other proteins in order to simultaneously integrate the diverse functions of transcription cofactors. We found an up-regulation of p300 expression in the AT1R TG in parallel with the U-STAT3. The increased formation of p300/U-STAT3 complex in the TG suggests that their association may be a hypertrophy-sensitive event. The report that p300 interacts with unphosphorylated-STAT6 to regulate cyclooxygenase-2 provided further support for our observation.36 STAT3-dependent recruitment of p300/CBP to junB promoter requires phosphorylation at Ser-727 in addition to the Tyr-705.12,37 However, we were able to exclude the involvement of phosphorylated Ser-727 of STAT3 in our experiments. We also excluded the possible influence of other STATs that can heterodimerize with STAT3 to modify function (data not shown).
Myocardial expression of OPN, a secreted soluble protein, was linked to LVH and HF.19 Its induction is inhibited by angiotensin-converting enzyme inhibitor, suggesting the involvement of Ang II in OPN gene regulation.38 Although not investigated here, CTGF is also a known hypertrophic factor in cardiac myocytes.39 Both CTGF and OPN are secreted soluble proteins that are potent cell type specific growth factors that could independently activate U-STAT3, forming a positive feedback loop in cardimyocytes.40 The expression of AT1R is known to mediate cardiac fibrosis associated with cardiomyocyte hypertrophy. The cues for the infiltration of fibroblasts and the deposition of extracellular matrix may be provided by the compromised myocytes in the form of secreted growth factors, like OPN, CTGF, and TGFβ, all of which may be U-STAT3 regulated in the chronic phase. Since the expression of predicted target genes was modulated by U-STAT3 in HEK cells, mechanistically this novel U-STAT3 transcription program could account for remodelling associated with HF.
RGS2 is a member of a large family of proteins that accelerate the termination of G protein-mediated signals and block signal generation in some instances.33 Cardiomyocyte growth and function are regulated by heterotrimeric G protein signals, fine tuning of which might be impaired by a reduction of RGS2 in the TG mice, a known risk in HF.20 Further studies are needed to clarify the mechanism of differential regulation of RGS2 by U-STAT3 in different species and/or cell types.
In summary, the present study suggests that U-STAT3 is an important transcription factor in the pathogenesis of HF induced by chronic AT1R activation. U-STAT3 is able to activate gene expression in response to Ang II and is able to regulate pathological processes such as AT1R-mediated cardiac hypertrophy and HF. We cannot exclude the possibility that overexpressed U-STAT3 inhibited the ‘classical’ function of phosphorylated STAT3, as reported by others,34 but the evidence presented indicates a novel gene regulatory mechanism mediated by U-STATs. The proposed U-STAT3 mechanism must interact with multiple signalling pathways conventionally implicated in cardiac remodelling. The animal and cellular models described in this study could thus help in the evaluation of U-STAT3-directed drug therapy strategies for reversal of hypertrophy. Our study indicates that consideration must be expanded to encompass U-STATs in describing molecular mechanisms of cardiac hypertrophy and to comprise accumulation of U-STAT3 as a prognostic marker for the disease.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by National Institutes of Health Grants R01 HL57470, HL083243 (to S.S.K.) and by American Heart Association Postdoctoral Fellowship Award #0825564D (to H.Y.).
Supplementary Material
Acknowledgements
We thank Drs Mona Nemer and Pierre Paradis (University of Montreal, Canada) for providing AT1R TG, Drs George R. Stark and Jinbo Yang for providing the U-STAT3 gene array data.
Conflict of interest: none declared.
References
- 1.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molkentin JD, Dorn GW., II Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 3.Barrios V, Calderon A, Escobar C, Barrios S, Navarro-Cid J, Gonzalez-Pedel V, et al. Electrocardiographic left ventricular hypertrophy regression induced by an angiotensin receptor blocker-based regimen in daily clinical practice: the SARA study. J Hypertens. 2007;25:1967–1973. doi: 10.1097/HJH.0b013e3282257145. [DOI] [PubMed] [Google Scholar]
- 4.Okin PM, Devereux RB, Jern S, Kjeldsen SE, Julius S, Nieminen MS, et al. Regression of electrocardiographic left ventricular hypertrophy by losartan versus atenolol: the Losartan Intervention for Endpoint reduction in Hypertension (LIFE) study. Circulation. 2003;108:684–690. doi: 10.1161/01.CIR.0000083724.28630.C3. [DOI] [PubMed] [Google Scholar]
- 5.Zou Y, Akazawa H, Qin Y, Sano M, Takano H, Minamino T, et al. Mechanical stress activates angiotensin II type 1 receptor without the involvement of angiotensin II. Nat Cell Biol. 2004;6:499–506. doi: 10.1038/ncb1137. [DOI] [PubMed] [Google Scholar]
- 6.Gallagher AM, Bahnson TD, Yu H, Kim NN, Printz MP. Species variability in angiotensin receptor expression by cultured cardiac fibroblasts and the infarcted heart. Am J Physiol. 1998;274:H801–H809. doi: 10.1152/ajpheart.1998.274.3.H801. [DOI] [PubMed] [Google Scholar]
- 7.Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am J Physiol Cell Physiol. 2007;292:C82–C97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 8.Tsai CT, Lai LP, Kuo KT, Hwang JJ, Hsieh CS, Hsu KL, et al. Angiotensin II activates signal transducer and activators of transcription 3 via Rac1 in atrial myocytes and fibroblasts: implication for the therapeutic effect of statin in atrial structural remodeling. Circulation. 2008;117:344–355. doi: 10.1161/CIRCULATIONAHA.107.695346. [DOI] [PubMed] [Google Scholar]
- 9.El-Adawi H, Deng L, Tramontano A, Smith S, Mascareno E, Ganguly K, et al. The functional role of the JAK-STAT pathway in post-infarction remodeling. Cardiovasc Res. 2003;57:129–138. doi: 10.1016/s0008-6363(02)00614-4. [DOI] [PubMed] [Google Scholar]
- 10.Leonard WJ, O'Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 11.Wen Z, Darnell JE., Jr Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 15.Paradis P, Dali-Youcef N, Paradis FW, Thibault G, Nemer M. Overexpression of angiotensin II type I receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proc Natl Acad Sci USA. 2000;97:931–936. doi: 10.1073/pnas.97.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramchandran R, Takezako T, Saad Y, Stull L, Fink B, Yamada H, et al. Angiotensinergic stimulation of vascular endothelium in mice causes hypotension, bradycardia, and attenuated angiotensin response. Proc Natl Acad Sci USA. 2006;103:19087–19092. doi: 10.1073/pnas.0602715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Tanaka K, Morioka K, Uesaka T, Yamada N, Takamori A, et al. Thymidine phosphorylase gene transfer inhibits vascular smooth muscle cell proliferation by upregulating heme oxygenase-1 and p27KIP1. Arterioscler Thromb Vasc Biol. 2005;25:1370–1375. doi: 10.1161/01.ATV.0000168914.85107.64. [DOI] [PubMed] [Google Scholar]
- 18.Yue H, Uzui H, Shimizu H, Nakano A, Mitsuke Y, Ueda T, et al. Different effects of calcium channel blockers on matrix metalloproteinase-2 expression in cultured rat cardiac fibroblasts. J Cardiovasc Pharmacol. 2004;44:223–230. doi: 10.1097/00005344-200408000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg M, Zugck C, Nelles M, Juenger C, Frank D, Remppis A, et al. Osteopontin, a new prognostic biomarker in patients with chronic heart failure. Circ Heart Fail. 2008;1:43–49. doi: 10.1161/CIRCHEARTFAILURE.107.746172. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Anger T, Su J, Hao J, Xu X, Zhu M, et al. Selective loss of fine tuning of Gq/11 signaling by RGS2 protein exacerbates cardiomyocyte hypertrophy. J Biol Chem. 2006;281:5811–5820. doi: 10.1074/jbc.M507871200. [DOI] [PubMed] [Google Scholar]
- 21.Ziesche E, Bachmann M, Kleinert H, Pfeilschifter J, Muhl H. The interleukin-22/STAT3 pathway potentiates expression of inducible nitric-oxide synthase in human colon carcinoma cells. J Biol Chem. 2007;282:16006–16015. doi: 10.1074/jbc.M611040200. [DOI] [PubMed] [Google Scholar]
- 22.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, et al. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 23.Ainscough JF, Drinkhill MJ, Sedo A, Turner NA, Brooke DA, Balmforth AJ, et al. Angiotensin II type-1 receptor activation in the adult heart causes blood pressure-independent hypertrophy and cardiac dysfunction. Cardiovasc Res. 2009;81:592–600. doi: 10.1093/cvr/cvn230. [DOI] [PubMed] [Google Scholar]
- 24.Reudelhuber TL, Bernstein KE, Delafontaine P. Is angiotensin II a direct mediator of left ventricular hypertrophy? Time for another look. Hypertension. 2007;49:1196–1201. doi: 10.1161/HYPERTENSIONAHA.106.075085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunisada K, Negoro S, Tone E, Funamoto M, Osugi T, Yamada S, et al. Signal transducer and activator of transcription 3 in the heart transduces not only a hypertrophic signal but a protective signal against doxorubicin-induced cardiomyopathy. Proc Natl Acad Sci USA. 2000;97:315–319. doi: 10.1073/pnas.97.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichihara S, Senbonmatsu T, Price E, Jr, Ichiki T, Gaffney FA, Inagami T. Angiotensin II type 2 receptor is essential for left ventricular hypertrophy and cardiac fibrosis in chronic angiotensin II-induced hypertension. Circulation. 2001;104:346–351. doi: 10.1161/01.cir.104.3.346. [DOI] [PubMed] [Google Scholar]
- 27.Ghatpande S, Goswami S, Mascareno E, Siddiqui MA. Signal transduction and transcriptional adaptation in embryonic heart development and during myocardial hypertrophy. Mol Cell Biochem. 1999;196:93–97. [PubMed] [Google Scholar]
- 28.Mascareno E, El-Shafei M, Maulik N, Sato M, Guo Y, Das DK, et al. JAK/STAT signaling is associated with cardiac dysfunction during ischemia and reperfusion. Circulation. 2001;104:325–329. doi: 10.1161/01.cir.104.3.325. [DOI] [PubMed] [Google Scholar]
- 29.Hunyady L, Catt KJ. Pleiotropic AT1 receptor signaling pathways mediating physiological and pathogenic actions of angiotensin II. Mol Endocrinol. 2006;20:953–970. doi: 10.1210/me.2004-0536. [DOI] [PubMed] [Google Scholar]
- 30.Sayeski PP, Ali MS, Frank SJ, Bernstein KE. The angiotensin II-dependent nuclear translocation of Stat1 is mediated by the Jak2 protein motif 231YRFRR. J Biol Chem. 2001;276:10556–10563. doi: 10.1074/jbc.M008856200. [DOI] [PubMed] [Google Scholar]
- 31.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci USA. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 33.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida Y, Kumar A, Koyama Y, Peng H, Arman A, Boch JA, et al. Interleukin 1 activates STAT3/nuclear factor-kappaB cross-talk via a unique TRAF6- and p65-dependent mechanism. J Biol Chem. 2004;279:1768–1776. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui X, Zhang L, Luo J, Rajasekaran A, Hazra S, Cacalano N, et al. Unphosphorylated STAT6 contributes to constitutive cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene. 2007;26:4253–4260. doi: 10.1038/sj.onc.1210222. [DOI] [PubMed] [Google Scholar]
- 37.Zhao H, Nakajima R, Kunimoto H, Sasaki T, Kojima H, Nakajima K. Region 752–761 of STAT3 is critical for SRC-1 recruitment and Ser727 phosphorylation. Biochem Biophys Res Commun. 2004;325:541–548. doi: 10.1016/j.bbrc.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 38.Graf K, Do YS, Ashizawa N, Meehan WP, Giachelli CM, Marboe CC, et al. Myocardial osteopontin expression is associated with left ventricular hypertrophy. Circulation. 1997;96:3063–3071. doi: 10.1161/01.cir.96.9.3063. [DOI] [PubMed] [Google Scholar]
- 39.Hayata N, Fujio Y, Yamamoto Y, Iwakura T, Obana M, Takai M, et al. Connective tissue growth factor induces cardiac hypertrophy through Akt signaling. Biochem Biophys Res Commun. 2008;370:274–278. doi: 10.1016/j.bbrc.2008.03.100. [DOI] [PubMed] [Google Scholar]
- 40.Ehrchen J, Heuer H, Sigmund R, Schafer MK, Bauer K. Expression and regulation of osteopontin and connective tissue growth factor transcripts in rat anterior pituitary. J Endocrinol. 2001;169:87–96. doi: 10.1677/joe.0.1690087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.