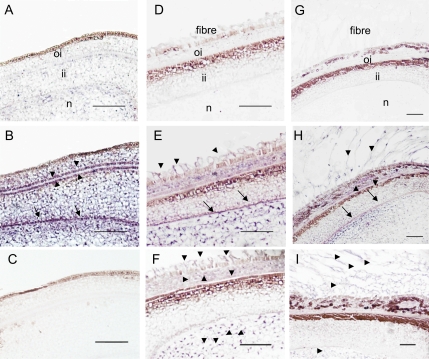
Fig. 6.
In situ mRNA localization of GhPEPC1 and 2 in cotton ovules and seeds. (A) Cross-section of ovules at –2 DAA hybridized with a GhPEPC1 sense probe as a negative control. (B) Cross-section of ovules at –2 DAA hybridized with a GhPEPC1 anti-sense probe. Note the absence of GhPEPC1 mRNA signals in the ovule epidermis but strong mRNA signals, indicated by the purple colour, in integument and nucellus, particularly at the interface between outer- and inner-integument (arrow heads) and at the innermost layer of inner integument (arrows). (C) Cross section of ovules at –2 DAA hybridized with a GhPEPC2 anti-sense probe. Note, no GhPEPC2 mRNA signals were detected. (D) Cross-section of seed at 1 DAA, hybridized with a sense probe of GhPEPC1. (E) Cross-section of seed at 1 DAA, hybridized with an antisense probe of GhPEPC1, showing the presence of GhPEPC1 mRNA in elongating fibres (arrows) in addition to that in the seed coat and nucellus. (F) Cross-section of seed at 1 DAA, hybridized with an antisense probe of GhPEPC2, showing the presence of its mRNA in elongating fibres in addition to that in the outer seed coat and nucellus (arrow heads). (G) Cross-section of seed at 5 DAA, hybridized with a sense probe of GhPEPC1. (H) Cross-section of seed at 5 DAA, hybridized with an anti-sense probe GhPEPC1. (I) Cross-section of seed at 5 DAA, hybridized with an antisense probe of GhPEPC2. oi, Outer integument; ii, inner integument; n, nucellus. Bars=50 μm.