Abstract
RING (really interesting new gene)-H2 domain-containing proteins are widely represented in plants and play important roles in the regulation of many developmental processes as well as in plant–environment interactions. In the present report, experiments were performed to unravel the role of the poplar gene PtaRHE1, coding for a RING-H2 protein. In vitro ubiquitination assays indicate a functional E3 ligase activity for PtaRHE1 with the specific E2 ubiquitin-conjugating enzyme UbcH5a. The overexpression of PtaRHE1 in tobacco resulted in a pleiotropic phenotype characterized by a curling of the leaves, the formation of necrotic lesions on leaf blades, growth retardation, and a delay in floral transition. The plant gene expression response to PtaRHE1 overexpression provided evidence for the up-regulation of defence- and/or programmed cell death-related genes. Moreover, genes coding for WRKY transcription factors as well as for mitogen-activated protein kinases, such as wound-induced protein kinase (WIPK), were also found to be induced in the transgenic lines as compared with the wild type. In addition, histochemical β-glucuronidase staining showed that the PtaRHE1 promoter is induced by plant pathogens and by elicitors such as salicylic acid and cellulase. Taken together, these results suggest that the E3 ligase PtaRHE1 plays a role in the ubiquitination-mediated regulation of defence response, possibly by acting upstream of WIPK and/or in the activation of WRKY factors.
Keywords: Defence response, E3 ligase, Nicotiana tabacum, Populus tremula×P. alba, RING-H2
Introduction
The poplar gene PtaRHE1, coding for a RING (really interesting new gene)-H2 domain-containing protein, has been identified through a cDNA-amplified fragment length polymorphism (AFLP) screening aimed at discovering genes whose expression is differential between the top and the base of 6-month-old Populus tremula×P. alba stems (van Raemdonck et al., 2005). In situ RT-PCR localization of PtaRHE1 in poplar stems undergoing secondary growth showed that this gene is mainly expressed within the cambial zone and, more particularly, in ray initials and derivatives (van Raemdonck et al., 2005). RING domains are characterized by four pairs of conserved cysteine (C) and histidine (H) residues coordinating two zinc ions in a cross-brace structure. Depending on the nature of metal ligands, but also on the spacing between pairs of metal ligands, RINGs have been classified into two main families, namely RING-HC (C3HC4) and RING-H2 (C3H2C3), and into other minor RING variants (Saurin et al., 1996; Jensen et al., 1998; Stone et al., 2005). As described by van Raemdonck et al. (2005) and in Supplementary Fig. S1 available at JXB online, the closest homologue (54% identity) to PtaRHE1 is the Arabidopsis thaliana ATL2 (Arabidopsis Toxicos en Levadura) (At3g16720) (Martínez-García et al., 1996) whose family members are characterized by a transmembrane (TM) domain, a basic domain, a conserved domain, a RING-H2 domain, and a highly divergent region in the C-terminal part of the protein (Serrano et al., 2006). As described by Serrano et al. (2006), the two T-DNA insertional mutants of ATL2 are not knock-out mutants and show no phenotype, and unfortunately cannot be used for complementation.
There is well-documented evidence showing that many plant RING domain-containing proteins act as E3 ubiquitin (Ub) ligases by promoting ubiquitination of specific target proteins. Ub attachment can be accomplished in different ways (including protein monoubiquitination, multiple monoubiquitination, and polyubiquitination) that determine the target's fate (Haglund and Dikic, 2005). The ubiquitination of protein targets requires the successive activity of the Ub-activating enzyme (E1), the Ub-conjugating enzyme (E2), and the Ub ligase (E3) which confers specificity to the degradation process (Schwechheimer et al., 2009). Monoubiquitination is considered to function as a regulatory signal that can mediate the activity, subcellular localization, or conformation of a protein (Haglund and Dikic, 2005). Polyubiquitination produced by the linkage of Ub to K48 of another Ub moiety is known as a signal for proteosomal degradation of modified target proteins via the Ub–26S proteasome pathway (Bachmair et al., 2001; Vierstra, 2003; Moon et al., 2004). The E3 ligase activity of 64 recombinant RING-containing A. thaliana proteins has been investigated by in vitro ubiquitination assays (Stone et al., 2005). Although >70% of these RING proteins were capable of mediating polyubiquitination in vitro, using AtUBC8 as E2 or other A. thaliana E2s from different subfamilies, 17 RING-H2 proteins were not, possibly as a consequence of misfolding in the expression host, or requirements for specific cofactors or E2 partners (Kraft et al., 2005; Stone et al., 2005).
The data reported in the literature indicate that RING proteins are associated with plant growth and development as well as with plant–environment interactions (Schwechheimer et al., 2009). For instance, in Arabidopsis, COP1 is involved in the repression of photomorphogenesis (von Arnim and Deng, 1994; Subramanian et al., 2004), BIG BROTHER in organ size by restricting the duration of cell proliferative growth (Disch et al., 2006), RHF1a and RHF2a in the formation of male and female gametophytes (Liu et al., 2008a), HUB1 and 2 in the control of cellular development during leaf and root development (Fleury et al., 2007) and in flowering time control (Cao et al., 2008), SHA1 in shoot apical meristem maintenance (Sonoda et al., 2007), XBAT32 in lateral root development (Nodzon et al., 2004), and RIE1 in seed development (Xu and Li, 2003), and in rice, EL5 was associated with root development (Koiwai et al., 2007).
Other RING finger proteins are involved in the regulation of hormone signalling pathways in A. thaliana, such as AIP2 (Zhang et al., 2005), KEG (Stone et al., 2006), and SDIR1 (Zhang et al., 2007) in abscisic acid (ABA) signalling, SINAT5 in auxin response (Xie et al., 2002), and BRH1 in brassinosteroid signalling (Molnár et al., 2002). In addition, RING proteins have been shown to regulate the response to biotic and abiotic stresses as well as to be involved in plant defence (Craig et al., 2009). For instance, RIN2 and RIN3 are involved in the RPM1- and RPS2-dependent hypersensitive response (HR) (Kawasaki et al., 2005), BAH1/NLA in the regulation of salicylic acid (SA) accumulation (Yaeno and Iba, 2008), RING1 in the triggering of the programmed cell death (PCD) pathway (Lin et al., 2008), XERICO in the regulation of drought tolerance through alteration of the ABA signalling pathway (Ko et al., 2006), and HOS1 in regulating cold responses (Lee et al., 2001).
Here, it is shown that the recombinant PtaRHE1 protein is a functional E3 ligase as demonstrated by its autoubiquitination. To characterize further the role of PtaRHE1, its overexpression was investigated. Instead of the poplar model, heterelogous expression in the tobacco model plant was chosen since tobacco produces typical angiosperm wood and homozygous lines can be rapidly obtained. Arabidopsis was not selected in this study since although it produces secondary xylem, it lacks ray parenchyma cells (Chaffey et al., 2002) where PtaRHE1 was found to be precisely expressed (van Raemdonck et al., 2005). Overexpressing PtaRHE1 resulted in dramatic alterations of leaf phenotype as well as in up-regulation of defence genes and genes encoding WRKY transcription factors. Challenging transgenic tobacco plants with different stresses showed that the PtaRHE1 promoter is responsive to several plant pathogens and to cellulase (Cel), as well as to ABA and SA. All together, these data suggest that PtaRHE1 might be part of the overall signal cascades involved in plant defence and development.
Materials and methods
Plant material and growth conditions
Non-transgenic and transgenic tobacco plants (Nicotiana tabacum cv. Havana) were grown aseptically on Murashige and Skoog (MS) medium (Micro and 1/2 concentration Macro elements including vitamins; Duchefa) supplemented with 200 mg l−1 kanamycin (Duchefa) when needed. Cultures were incubated at 23±2 °C under a 16 h light photoperiod (70 μmol m−2 s−1, cool-white fluorescent lamp; Osram). Sown seeds, or acclimatized plants, were cultivated on soil in a growth chamber under a 16 h light photoperiod at 24 °C.
Plant treatment
For biotic stress treatment, 19-day-old pPtaRHE1::GUS plantlets grown on phytagel (0.2%, w/v) solidified MS medium were inoculated in 20 ml of liquid MS medium containing Rhodococcus fascians (strain D188), Pseudomonas syringae pv tabaci, or Agrobacterium tumefaciens (strain C58) (500 μl of overnight bacterial culture in 2 ml of liquid YEB medium).
For abiotic stress treatment, 12-day-old pPtaRHE1::GUS plantlets grown in solid MS medium were transferred to fresh liquid MS medium containing ABA (150 μM), H2O2 (10 mM), SA (50 μM), NaCl (300 mM), Cel (100 μg ml–l), spermidine (0.5 mM), or spermine (0.5 mM). For each treatment, seedlings were harvested after 8 h for β-glucuronidase (GUS) staining.
Vector construction for PtaRHE1 overexpression and plant transformation
For overexpression, the coding sequence of PtaRHE1 (AY780430), cloned in pCR®4-TOPO® (Invitrogen, Merebelke, Belgium), was amplified with the primer attb1RHE1 5′-AAAAAGCAGGCTTAATGGACCCAGACTCG-3′ to flank the attB1 recombination site at the 5′ end of the coding sequence and the primer 5′-AGAAAGCTGGGTCTTAACACCGAGTTTGC-3′ to flank the attB2 recombination site to the 3′ end of the sequence with the stop codon. A second PCR using primers amplifying the entire attB1 and attB2 sequences was performed, according to the supplier's instructions (Invitrogen). The PCR fragment was cloned in the Gateway™ vector pDONR221 yielding the entry clone RING BP2, which was then recombined with the Gateway™-compatible T-DNA destination vector pK7WG2 containing a caulifower mosaic virus (CaMV) 35S promoter (Karimi et al., 2002), in reactions mediated by the Gateway™ BP and LR Clonase™ Enzyme Mix (Invitrogen).
The resulting RLR1 construct was mobilized to Agrobacterium strain C58C1Rif containing the plasmid pGV2260. N. tabacum was transformed by the leaf disc protocol according to Deblaere et al. (1987), using thidiazuron (1 mg l−1) instead of benzylaminopurine. The number of T-DNA inserts was assessed by segregation of T0 offspring on selective medium (MS supplemented with 200 μg ml−1 kanamycin). Eight T1 seedlings of each one-copy line were grown in the greenhouse and their seeds were sown on selective medium to identify homozygous lines (T2).
Semi-quantitative RT-PCR and real-time quantitative RT-PCR (RT-qPCR) analyses
Total RNA from leaves of 6-week-old plants was prepared using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) then treated with DNase I (DNA-free™ from Ambion, Austin, TX, USA). RNA quality and quantity were assessed with a Bioanalyzer 2100 (Agilent). Single-stranded cDNA was synthesized using the Reverse Transcription System (Promega, Madison, WI, USA). For semi-quantitative PCR, reactions were performed using Promega's MasterMix. Expression levels of PtaRHE1 and EF1α were assessed by means of RT-PCR using the primers as described in Supplementary Table S1 at JXB online.
RT-qPCR analysis was performed as described by Vandeputte et al. (2007), in an ABI 7900 system (Applied Biosystems). Transcriptional changes were calculated based on the comparative ΔΔCT method as described by Livak and Schmittgen (2001) and are reported as ratios between expression in transgenic lines overexpressing PtaRHE1 (RLR1-1-1 and RLR1-5-7) and wild-type (WT) plants. The CT value of each gene was normalized to the CT value of the reference gene EF1α. The expression of each gene was investigated in three biological replicates. Primer pairs used for RT-PCR analysis were designed according to the cDNA sequences present in public databases (Supplementary Table S1 at JXB online). Criteria for designing primers (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) were a primer size between 18 and 25, an optimal Tm at 60 °C, and a product size ranging from 200 bp to 250 bp.
PtaRHE1 promoter cloning and analysis
Genomic DNA was extracted from P. tremula×P. alba (clone INRA 717-1B4) aerial parts using a DNeasy Plant Mini Kit (Qiagen). pPtaRHE1 was recovered using a GenomeWalker™ Kit (Clontech), according to the manufacturer's instructions. Several rounds of genome walking were performed with the following gene-specific primers: GSP1 5′-GTTACTTACTCATCTAACCGGGTCAAG-3′, GSP2 5′-ACAGTACCTCTTCTTTCCCTACTTAGC-3′, GSP1 5′-TCTCGAGTCTGGGTCCATTTCTTGAAT-3′, GSP1 5′-AGAGGAGGAGGACGAGGTAGGTTCTTG-3′, and GSP2 5′-TGATCAAACCAATTCACCTTCCTCTCA-3′. The promoter sequence was amplified with primers F 5′-CAAGTTGCAACCGGATTATG-3′ and R 5′-TTCAATTGGTGGATCTCTCG-3′, cloned in the pCR®4-TOPO® vector (Invitrogen), and sequenced. Prediction of potential cis-elements was performed using the PLACE database (Higo et al., 1999; http://www.dna.affrc.go.jp/htdocs/PLACE). Gateway® (Invitrogen) attB1 and attB2 sequence extensions were added for cloning into the Gateway®-compatible binary T-DNA destination vector pKGWFS7 (Karimi et al., 2002) allowing the fusion of the PtaRHE1 promoter with both GFP (green fluorescent protein) and GUS reporter genes in reactions mediated by the Gateway® BP and LR clonase® Enzyme mix (Invitrogen). Transgenic tobacco plants were produced as described above. Five homozygous lines were identified as having a similar pattern of expression during seedling development. One of these lines, RLR6-11-6, was selected for detailed expression analysis. Histochemical GUS staining was performed as described by Hemerly et al. (1993). Seedlings were examined under a light binocular (Olympus SZX-ILLK200), and roots and cross-sections under a light microscope (Olympus BX 60). Images were acquired with a Colorview II Soft Imaging System (Olympus).
Production of PtaRHE1 and PtaRHE1-Ct recombinant proteins
To produce the full-length protein, PtaRHE1 was amplified by PCR (1 min at 94 °C, 1 min at 60 °C, 1 min 30 s at 68 °C for 30 cycles, followed by 10 min at 68 °C) with primers F 5′-AAAAAGCAGGCTTAATGCAGAAAGAAAAA-3′, and R 5′-AGAAAGCTGGGTATAGATAAAAGGCATA-3′. To produce PtaRHE1 where the N-terminal TM and the basic domains were deleted (PtaRHE1-Ct), the same coding sequence was amplified by PCR (1 min at 94 °C, 1 min at 50 °C, 1 min 30 s at 68 °C for 30 cycles, followed by 10 min at 68 °C) with the primers F 5′-AAAAAGCAGGCTACCACATGCCAGATTCT-3′ and R 5′- AGAAAGCTGGGTATAGATAAAAGGCATA-3′. PCRs were performed using the Platinum Pfx polymerase (Invitrogen). The pBAD-DEST49 expression system (Invitrogen) was used to produce recombinant proteins with horseradish peroxidase (HRP)–thioredoxin as an N-terminal fusion partner (14 kDa) of the cloned gene product, a V5 epitope, and a hexahistidine (6 His) tag (4 kDa) as C-terminal fusion partners, resulting in fusion proteins with an expected mass of 50.9 kDa for PtaRHE1 and 42.6 kDa for PtaRHE1-Ct.
In vitro autoubiquitination assay
PtaRHE1 and PtaRHE1-Ct were expressed in the Escherichia coli strain TOP10 (Invitrogen) for recombinant protein production and purification. Protein production was induced with 0.02% arabinose for 3 h at 37 °C. After pellet lysis, the PtaRHE1-His and PtaRHE1-Ct-His proteins were bound to Ni-NTA beads (Invitrogen), washed, but not eluted. The proteins were allowed to refold in a buffer containing zinc ions (20 nM HEPES pH 7.4, 150 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.05% Triton X-100, 0.02 mM ZnCl2, 1.5 mM MgCl2). For the in vitro autoubiquitination assay, ATP-containing buffer [50 mM HEPES pH 7.4, 2 mM ATP, 5 mM MgCl2, 2 mM dithiothreitol (DTT), 0.2 mM ZnCl2], haemagglutinin (HA)-tagged Ub (human recombinant), E1 Ub-activating enzyme (rabbit recombinant), and several human recombinant E2 Ub-conjugating enzymes, including UbcH5a, UbcH5b and UbcH5c, purchased from BostonBiochem, were used as reagents. Ubiquitination reactions including negative controls for E3, E2 and Ub were incubated at 30 °C for 1 h. The reactions were stopped by adding SDS loading buffer and incubation at 65 °C for 10 min. The samples were subjected to 8% SDS–PAGE and blotted on Immobilon®-P polyvinylidene difluoride (PVDF) membranes (Sigma). The PtaRHE1-His and PtaRHE1-Ct-His proteins were detected by penta/tetra His antibody (Qiagen) and Ub or the ubiquitinated proteins by anti-HA antibody (Roche) against HA-Ub.
Accession number
Sequence data for the PtaRHE1 promoter region can be found in the GenBank/EMBL data libraries under the accession number GQ174438.
Results
In vitro ubiquitination assays indicate that PtaRHE1 is a functional E3 ligase
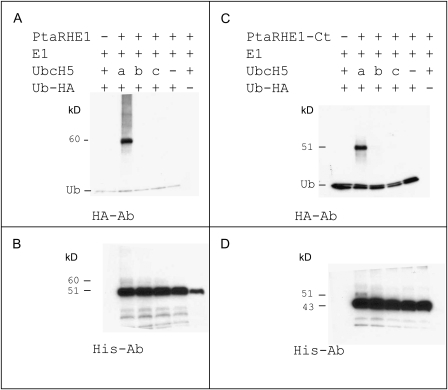
In order to investigate the enzymatic activity of PtaRHE1, recombinant PtaRHE1 proteins were produced in E.coli and purified for in vitro autoubiquitination assays. Since PtaRHE1 possesses a putative N-terminal TM domain that may interfere with its production in E.coli (as reported for EL5 by Takai et al., 2002), two recombinant forms of PtaRHE1 were produced. The first form was the full-length protein (PtaRHE1) and the second form was a truncated protein (PtaRHE1-Ct) where the TM and the basic domains were deleted (Supplementary Fig. S2 at JXB online). The His-tagged PtaRHE1 and PtaRHE1-Ct proteins were purified and subjected to autoubiquitination in the presence of ATP, HA-tagged Ub (human recombinant), E1 (rabbit recombinant), and several human recombinant E2s, including UbcH5a, UbcH5b, and UbcH5c. After completion, these reactions were separated by SDS–PAGE and transferred to a PVDF membrane for immunodetection of the PtaRHE1 and PtaRHE1-Ct proteins and HA-Ub. The overlaps of either PtaRHE1 or PtaRHE1-Ct and HA-Ub signals on the membranes were verified to determine whether these proteins were coupled to Ub or not. As shown in Fig. 1A, following immunostaining with the anti-HA antibody, a band with a mol. wt of 60.3 kDa, corresponding to a shift of PtaRHE1 by 9.4 kDa (corresponding to the molecular weight of one Ub moiety), was observed only when UbcH5a was used as E2. A comparable pattern, albeit weaker, was obtained using the anti-His antibody against PtaRHE1 (Fig. 1B). These data clearly show that PtaRHE1 possesses an E3 ligase activity. Similarly to EL5 (Takai et al., 2002), PtaRHE1 uses specific E2 enzymes, in the present case UbcH5a, while UbcH5b and UbcH5c were not able to mediate the ubiquitination reaction. The truncated PtaRHE1-Ct also showed a preference for UbcH5a. However, in this case, only monoubiquitination patterns could be detected in the overlapping HA and His signals (Fig. 1C, D, respectively), suggesting that PtaRHE1-Ct might have a reduced activity and therefore that the deleted TM domain or areas near to it play a role in the PtaRHE1 ubiquitination activities. Hetero- or homodimerization has been shown to be essential for the function of many E3 ligases (Nikolay et al., 2004; Subramanian et al., 2004) and it could be that, due to the deletion, a dimerization has become impossible. Another possibility is that interactions with the other reaction components, such as E2, can not take place efficiently due to the deletion.
Fig. 1.
E3 Ub ligase activity of PtaRHE1 and PtaRHE1-Ct proteins. (A) and (B) E3 ligase activity of HRP–thioreodoxin–PtaRHE1-6His fusion protein. (C) and (D) E3 ligase activity of HRP–thioreodoxin–PtaRHE1-Ct-6His fusion protein. Anti-HA antibody was used to detect Ub and ubiquitinated proteins (A, C) and anti-His antibody was used to detect His-tagged PtaRHE1 and PtaRHE1-Ct (B, D). Three different E2s, UbcH5a–c, were tested in this assay.
Overexpression of PtaRHE1 in transgenic tobacco triggers leaf curling, leaf blade necrosis as well as growth retardation and flowering delay
To investigate the role of PtaRHE1, transgenic tobacco lines overexpressing the full-length PtaRHE1 coding sequence under the control of the CaMV 35S promoter were generated. Four independent T2 transgenic lines, designated RLR1-1-1, RLR1-5-7, RLR1-7-6, and RLR1-9-3, were selected. Overexpression of PtaRHE1 in these lines was confirmed by RT-PCR analysis, whereas no amplicon could be detected in the WT plant (Fig. 2). Among these lines, RLR1-1-1 and RLR1-5-7 were selected for further characterization. After 1 month of growth, transgenic lines showed a curling of the leaves and necrotic spots on leaf blades that were both absent in WT plants (Fig. 3A, B). The severity of the phenotype varied from plant to plant, and ranged from leaf curling of all leaves (RLR1-1-1) to leaf curling limited to the youngest leaves (RLR1-5-7). In addition, leaf development was strongly restricted in RLR1-1-1 as compared with RLR1-5-7 and the WT (Fig. 3B). Three-month-old transgenic lines showed an altered development as compared with the WT plants (Fig. 3C). Transgenic plants were shorter, mainly because they formed shorter internodes, and showed a delay in floral transition (Fig. 3C). After 4 months, PtaRHE1 overexpressors reached the same size as the WT plants and floral transition occurred (Fig. 3D). The phenotype of the 4-month-old line RLR1-1-1 was characterized by a pronounced leaf curling (Fig. 3E) and by the appearance of chlorosis and necrosis on the leaf blades (Fig. 3F), whereas the WT leaves were flat (Fig. 3G) and did not show any necrosis (Fig. 3H). No apparent anatomical differences were noticed in transversal stem cross-sections, the main vein in leaves, and stomata density and distribution between transgenic lines and the WT (data not shown). However, transverse sections in leaves showed that RLR1-1-1 leaves (Fig. 3I) were less thick than WT leaves (Fig. 3J), possibly because intercellular spaces within the spongy parenchyma in the PtaRHE1 overexpressors were smaller and less frequent than in the WT plants.
Fig. 2.
Identification of transgenic lines overexpressing PtaRHE1. RT-PCR analysis of PtaRHE1 expression in T2 transgenic tobacco lines and the WT. EF1α was used as loading control.
Fig. 3.
Phenotype of transgenic lines overexpressing PtaRHE1. (A) and (B) One-month-old plants and third leaves, respectively. (C) Three-month-old plants. (D) Four-month-old plants. (E) and (F) Details of 4-month-old RLR1-1-1 leaves and leaf blade, respectively. (G) and (H) Details of 4-month-old WT leaves and leaf blade, respectively. (I) and (J) Hand-made cross-section in third leaves of 4-month-old RLR1-1-1 and WT plants, respectively. The arrows indicate necrotic spots. *, 3-month-old WT plant undergoing floral transition.
Several classes of genes encoding inducible defence-related proteins are up-regulated in transgenic plants overexpressing PtaRHE1
Since the phenotype of the PtaRHE1 overexpressors, i.e. the formation of necrotic lesions, was reminiscent of possible cellular processes linked to cell defence and/or cell death, the expression of a number of genes reported to be induced during several associated mechanisms in plant cells was investigated. As shown in Table 1, the selected genes are classified into several categories including genes induced during defence and the HR, genes associated with cell death linked to proteasome malfunction, genes induced during apoptosis, or genes linked to age-mediated leaf senescence and oxidative stress. Specific primers, listed in Supplementary Table S1 at JXB online, were designed for these genes either using the tobacco sequences in the database or using the closest homologous sequences of N. tabacum by performing a BLAST N search in the non-redundant NCBI database. Their expression was analysed in leaves of 1-month-old T2 plants of RLR1-1-1, RLR1-5-7, and the WT, when the phenotype was clearly visible. A quantitative analysis of the expression of the selected genes was performed by RT-qPCR using the cDNA made from three individual plants of each line. The overexpression of PtaRHE1 in each individual plant was first checked by RT-PCR (Supplementary Fig. S3 at JXB online).
Table 1.
Relative expression (fold) of a selection of genes associated with various types of cell processes linked to defence and/or cell death in two lines of PtaRHE1 overexpressors compared with the WT
| 2e–ΔΔCT RLR1-1-1 |
2e–ΔΔCT RLR1-5-7 |
|||||
| Mean | SE | t-test (P value) | Mean | SE | t-test (P value) | |
| PtaRHE1 | 828 735.18 | 300 840.44 | ≤0.01 | 209 593.44 | 57 530.96 | ≤0.01 |
| 1. Defence/elicitor-inducible genes | ||||||
| PR-1a | 256 701.17 | 176 302.06 | ≤0.01 | 8458.53 | 6585.74 | ≤0.01 |
| PR-1b | 120 967.23 | 85 436.22 | ≤0.01 | 1604.42 | 689.56 | ≤0.01 |
| TIZZ | 61 481.94 | 33 817.36 | ≤0.01 | 532.02 | 296.37 | ≤0.01 |
| PR5 | 444.98 | 377.28 | ≤0.01 | 9.03 | 1.85 | ≤0.01 |
| SAR8.2a | 102.64 | 34.82 | ≤0.01 | 5.81 | 2.72 | ≤0.01 |
| PR3 | 52.36 | 24.10 | ≤0.01 | 2.80 | 0.53 | ≤0.01 |
| HSR201 | 22.92 | 7.78 | ≤0.01 | 0.57 | 0.10 | NSa |
| HMGR2 | 20.16 | 10.37 | ≤0.01 | 0.68 | 0.02 | ≤0.01a |
| WRKY12 | 7.45 | 5.02 | ≤0.01 | 0.97 | 0.36 | NSa |
| EDS1 | 9.05 | 3.52 | ≤0.01 | 1.25 | 0.29 | NSa |
| PR4 | 5.84 | 1.47 | ≤0.01 | 0.98 | 0.29 | NSa |
| WIPK | 2.43 | 0.57 | ≤0.01 | 0.99 | 0.03 | NSa |
| LSD1 | 1.93 | 0.35 | ≤0.01a | 0.84 | 0.39 | NSa |
| SAMDC | 1.59 | 0.35 | NSa | 0.65 | 0.10 | ≤0.01a |
| BECLIN1 | 1.59 | 0.57 | NSa | 1.20 | 0.57 | NSa |
| DND1 | 1.59 | 0.43 | NSa | 1.52 | 0.77 | NSa |
| ABAKIN | 1.49 | 0.55 | NSa | 0.87 | 0.20 | NSa |
| NPR1 | 1.43 | 0.24 | NSa | 1.31 | 0.21 | NSa |
| HSP90 | 1.17 | 0.56 | NSa | 0.77 | 0.12 | NSa |
| SIPK | 1.01 | 0.05 | NSa | 0.82 | 0.14 | NSa |
| S26-PR6 | 0.95 | 0.23 | NSa | 2.09 | 1.33 | NS |
| NtMEK2 | 0.91 | 0.07 | NSa | 0.68 | 0.14 | ≤0.05a |
| HSR203J | 0.78 | 0.15 | NSa | 0.52 | 0.08 | ≤0.01a |
| Spermidine synthase | 0.67 | 0.03 | NSa | 0.81 | 0.17 | NSa |
| WIZZ | 0.66 | 0.22 | NSa | 0.88 | 0.32 | NSa |
| NOA1 | 0.53 | 0.09 | ≤0.05a | 0.78 | 0.07 | NSa |
| Polyamine oxidase | 0.54 | 0.27 | NSa | 0.52 | 0.16 | ≤0.01a |
| PR1c | – | – | – | – | – | – |
| HSR515 | – | – | – | – | – | – |
| 2. Cell death induced by proteasome malfunction-related genes | ||||||
| 20S proteasome α subunit 3 | 1.34 | 0.26 | NSa | 1.38 | 0.54 | NSa |
| Ubiquitin protease 6 | 1.35 | 0.56 | NSa | 1.12 | 0.13 | NSa |
| Ubiquitin protease 12 | 1.03 | 0.15 | NSa | 1.38 | 0.44 | NSa |
| BS2 | 1.17 | 0.42 | NSa | 1.06 | 0.32 | NSa |
| NAM-like | 0.88 | 0.42 | NSa | 1.14 | 0.11 | NSa |
| ClpP | 0.61 | 0.17 | NSa | 0.81 | 0.10 | NSa |
| 3. Both defence and proteasome malfunction cell death-induced genes | ||||||
| PR2 | 21 585.51 | 18 447.37 | ≤0.01 | 1169.14 | 1116.96 | ≤0.01 |
| HIN1 | 10.89 | 5.67 | ≤0.05 | 0.46 | 0.40 | NS |
| NtCP-23 | 2.01 | 0.70 | NS | 1.54 | 0.57 | NSa |
| SGT1 | 2.16 | 0.71 | NS | 2.27 | 0.88 | NS |
| 4. Apoptosis-related genes | ||||||
| BI-1 | 1.71 | 0.46 | NSa | 0.93 | 0.36 | NSa |
| DAD1 | 1.19 | 0.32 | NSa | 1.07 | 0.48 | NSa |
| 5. Leaf senescence-specific gene | ||||||
| CP1 | – | – | – | – | – | – |
| 6. Oxidative stress-related genes | ||||||
| APX | 1.10 | 0.28 | NSa | 0.70 | 0.10 | ≤0.05a |
| SOD | 0.69 | 0.07 | NSa | 0.58 | 0.06 | ≤0.05a |
| NbrbohA | 2.32 | 0.98 | NS | 0.63 | 0.36 | NSa |
The quantitative expression level of each gene was measured by RT-qPCR and each value is the relative accumulation of each gene transcript compared with that of EF1α. The data presented are the mean value of RNA preparations in three different individual plants of each line.
–, no expression detected; NS, not significant.
Genes indicated in bold underwent a significant change in expression in RLR1-1-1.
A difference in expression lower than 2-fold up or 2-fold down was not considered in this study.
As shown in Table 1, the two transgenic lines robustly expressed PtaRHE1, and RLR1-1-1, showing the strongest phenotype (Fig. 3), had four times more transcripts than RLR1-5-7. In RLR1-1-1, several genes coding for defence-related proteins were significantly induced. The transcript of PR-1a dramatically accumulated, with a relative increase of 250 000-fold as compared with the WT. Other defence-related genes were also up-regulated, including PR-1b, TIZZ, PR5, SAR8.2.A, PR3, HSR201, HMGR2, WRKY12, EDS1, PR4, and WIPK. Transcript levels of the tested genes related to proteasome cell death were not significantly different in the WT and in the transgenic lines. Two genes, PR2 and HIN1, known to be induced during both the HR and proteasome cell death (Kim et al., 2003), were also induced in RLR1-1-1. No significant changes in the transcript levels of the genes associated with apoptosis, senescence, or oxidative stress were detected in this transgenic line as compared with the WT. In the second transgenic line, RLR1-5-7, only some of the genes that are affected in line RLR1-1-1 had a significant change in expression, including PR-1a, PR-1b, PR2, PR3, PR5, SAR8.2A, and TIZZ. The gene expression analysis suggests that at least some phenotypic features observed in the lines overexpressing PtaRHE1 seem to be associated with a defence-like response of the plants. To support this conclusion, in planta analysis of PtaRHE1 gene expression in response to various stresses and during plant development was examined.
PtaRHE1 promoter-driven GUS activity is induced by various biotic and abiotic treatments and is developmentally regulated
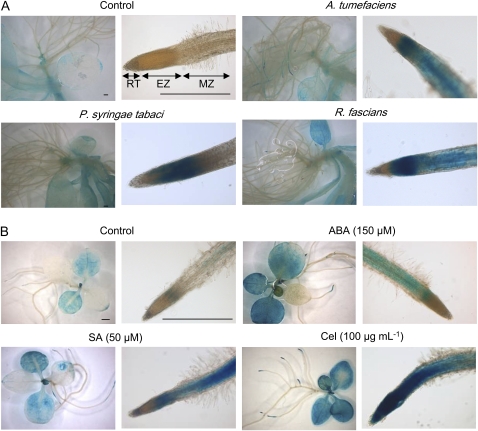
A 1207 bp long sequence upstream of the ATG codon of PtaRHE1 was isolated from P. tremula×P. alba by genome walking (Supplementary Fig. S4 at JXB online). The 74 bp 5′-untranslated region (UTR) initially recovered by rapid amplification of cDNA ends (RACE) (van Raemdonck et al., 2005) was extended to 102 bp by the homology with the expressed sequence tag (EST) Q044A08 from the contig POPLAR.9452.C1 (98% sequence similarity). Using the PLACE database, the PtaRHE1 promoter (pPtaRHE1) was searched for the presence of cis-regulatory sequences. As shown in Supplementary Fig. S3 at JXB online, several putative CAAT and TATA boxes were found. A number of potential cis-acting elements have also been identified including two ABA-responsive elements (ABREs), eight W-boxes, 10 ARR1AT elements, 11 GT-1-binding motifs, one BS1 site, 10 POLLENLELAT52 elements, three ACGTERD1 sequences, two ACGTABOX elements, six ROOTMOTIFTAPOX1 motifs, one RAV1AAT sequence, seven MYB recognition sites, nine GTGA motifs, eight NODCON2GM sequences, 12 DOF recognition sites, three MYC recognition sequences, and one HDZIP2ATATHB2 site. Although the function of these putative elements in the regulation of the expression of the PtaRHE1 gene remains to be elucidated, the analysis of the expression of the GUS gene driven by pPtaRHE1 upon biotic and abiotic stress and during development supported the function of some of the regulatory elements. Since a large number of putative stress-responsive elements are present in pPtaRHE1 (such as W-boxes, GT-1s, ABREs, ACGTERD1, and MYB elements; see Supplementary Fig. S4 at JXB online), pPtaRHE1::GUS plants were confronted with various biotic and abiotic treatments (see Materials and methods). As shown in Fig. 4A, although 19-day-old plantlets co-cultured for 24 h with different bacteria (A. tumefaciens, R. fascians, and P. syringae pv tabaci) displayed a similar GUS pattern in the aerial parts, a clear induction of pPtaRHE1 was visible in the root system. In the non-treated plants, GUS activity was not detected in the root, whereas following bacterial infection the promoter was clearly induced in the elongation and maturation zones of the root. To examine whether the response to pathogenic bacteria is linked to defence mechanisms, 12-day-old plants were treated with Cel and SA. Compared with the control, these two treatments (Cel in particular) strongly induced pPtaRHE1 even in the aerial parts (Fig. 4B). Moreover, ABA induces pPtaRHE1, perhaps due to the occurrence of ABREs in the promoter sequence. No obvious differences were observed for the other abiotic treatments (H2O2, NaCl, spermidine, or spermine; data not shown).
Fig. 4.
pPtaRHE1 response to various biotic and abiotic stresses. (A) Nineteen-day-old pPtaRHE1::GUS transgenic tobacco plants treated for 8 h with phytopathogens. (B) Twelve-day-old pPtaRHE1::GUS transgenic tobacco plants treated with various abiotic stresses. EZ elongation zone; MZ, maturation zone; RT, root tip. Scale bars represent 1 mm.
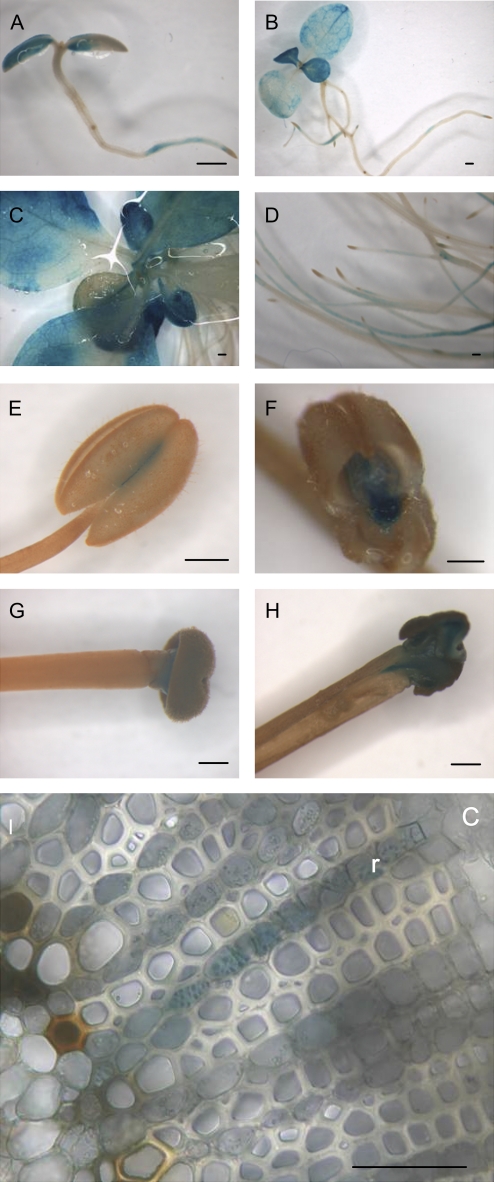
Histochemical GUS stainings were performed during plant development. As shown in Fig. 5A, 7 d post-germination, GUS activity was detected in the cotyledons and in the primary root but not within the root tips. A similar pattern of expression was observed in 12- and 33-day-old plantlets (Fig. 5B–D). In 3-month-old plants, GUS expression was also detected in anther, mainly in the degrading connective tissue, and in the stigma (Fig. 5E–H). A cross-section of the stem allowed the detection of GUS staining in ray parenchyma cells within the xylem (Fig. 5I). In conclusion, the expression of PtaRHE1 is associated with specific tissues or cell types, is developmentally regulated, and is induced by environmental factors.
Fig. 5.
pPtaRHE1-driven expression during plant development. (A) Seven-day-old seedling. (B) Twelve-day-old plant. (C) Aerial part of a 33-day-old plant. (D) Roots of a 33-day-old plant. (E–I) Three-month-old plants. (E) Anther. (F) Section in an anther showing expression in connective tissue. (G) Style. (H) Expression in the stigma. (I) Secondary xylem. c, cambium; r, ray. Scale bars represent 1 mm in A–H and 100 μm in I.
Discussion
Here, PtaRHE1, a poplar RING-containing protein, was shown to possess an E3 ligase activity since it is able to mediate its own ubiquitination in an in vitro assay (Fig. 1). The target substrate of PtaRHE1 has not yet been identified, but this E3 ligase was shown to function with the particular E2 UbcH5a. The interaction of ATL proteins, which are closely related to PtaRHE1, with members of the Ubc4/5 subfamily of E2s has already been reported. The rice EL5 functions in co-operation with UbcH5a and Ubc4, and their rice counterparts, OsUBC5a and OsUBC5b (Takai et al., 2002; Katoh et al., 2003, 2005). In accordance with this, most of the residues crucial for EL5–OsUBC5b binding, as identified by mutation analysis (Katoh et al., 2005), including V136, L138, D163, W165, L174, and R176, are conserved in the PtaRHE1 RING-H2 finger domain and correspond to V113, L115, D140, W142, L151, and R153 (Supplementary Fig. S2 at JXB online). Aguilar-Henonin et al. (2006) showed that the toxicity of the expression of A. thaliana ATL2 in Saccharomyces cerevisiae (Martínez-García et al., 1996) was alleviated when components of the yeast Ub–26S proteasome pathway were mutated, among others the E2 Ubc4. These data suggest that ATL2 also interacts with the Ubc4/5 E2 family to mediate ubiquitination of the target substrate. Performing in vitro ubiquitination assays with poplar homologues of these E2s could further validate the interaction of the Ubc4/5 subfamily with PtaRHE1.
Overexpressing PtaRHE1 in transgenic tobacco plants resulted in a marked alteration of plant development, characterized by a slower growth, a precocious inward curling of the leaves, the formation of necrotic lesions on leaf blades, and a delay in floral transition (Fig. 3). Increasing evidence supports a role for the Ub–proteasome system and protein degradation in plant development (Stone and Callis, 2007), in the regulation of PCD (Kim et al., 2003, 2006), and in plant defence against pathogens (reviewed by Zheng et al., 2006; Craig et al., 2009). The pleiotropic phenotype observed in PtaRHE1 overexpressors may therefore result from alterations of processes linked to ubiquitination. Alteration of plant development has been observed following the overexpression of several RING-H2 proteins belonging to the ATL family. For instance, the overexpression of MsRH2-1 in alfalfa and in A. thaliana caused a shortening of plant stature, increased apical dominance, leaf hyponasty, inhibition of leaf venation and lateral root development, delayed nodulation in the case of alfalfa, and abnormal flower development, probably due to a disruption of auxin signalling pathway(s) (Karlowski and Hirsh, 2003). In contrast, transgenic tobacco overexpressing OsBIRF1 were characterized by an increased growth and had more leaves than the WT plants (Liu et al., 2008b). Finally, the overexpression of EL5 in rice resulted in root growth arrest, due to the possible role of EL5 in the maintenance of cell viability after the initiation of root formation (Koiwai et al., 2007).
The phenotype described for PtaRHE1 tobacco overexpressors does not seem to be unique to RING-H2 proteins since similar phenotypes have also been reported in plants where other genes are up-regulated. For instance, the overexpression of the N. plumbaginifolia gene encoding the ankyrin repeat protein glucanohydrolase-binding protein 1 (GBP1) resulted in downward curling of leaves accompanied by necrotic lesions (Wirdnam et al., 2004). By performing grafting experiments, these authors reported that the signal inducing leaf curling was transmissible acropetally, suggesting that the sugar transport through the phloem was altered, with disturbances in carbohydrate metabolism leading to leaf curling as a consequence. In the case of PtaRHE1-overexpressing lines, grafting experiments did not show signal transmission and comparative analysis of carbohydrates did not reveal significant qualitative and quantitative differences between stems of WT and RLR1-5-7 lines (data not shown).
The HR-like cell death phenotype observed in leaves of PtaRHE1-overexpressing lines may be related to an alteration of the Ub–proteasome pathway. For instance, the overexpression of a variant form of Ub, where K48 is replaced by an R, inhibits proteolysis and induced a phenotype in tobacco similar to the one observed in PtaRHE1 overexpressors, characterized by shorter internode length, leaf curling, abnormalities in vascular tissues, and formation of necrotic lesions on leaves (Bachmair et al., 1990). In the present study, none of the selected genes specifically related to proteasome malfunction (Kim et al., 2003, 2006; Table 1) was up-regulated in PtaRHE1-overexpressing lines, suggesting that proteasome functioning is not affected in these lines. This hypothesis is supported by Kim et al. (2003, 2006) who showed that the silencing of two different subunits of the 26S proteasome, the α6 subunit of the 20S proteasome and the RPN9 subunit of the 19S regulatory complex, activated the PCD process. As shown by these authors, this proteasome-mediated cell death stimulated the expression of only a subset of transcripts that are highly induced during P. syringae pv syringae-mediated HR, indicating that diverse PCD pathways co-exist in plant cells with differential regulation mechanisms.
Numerous defence-related genes are strongly induced in PtaRHE1 overexpressors, suggesting a role for PtaRHE1 in defence mechanisms (Table 1). The most strongly increased gene in 35S::PtaRHE1 lines is PR-1a, which encodes an acidic protein widely represented in plants that is considered as a marker for systemic acquired resistance (SAR) and as the most abundant pathogenesis-related (PR) protein in infected plant tissue (Ward et al., 1991; van Loon et al., 2006). Other PR genes are up-regulated in PtaRHE1 overexpressors, including genes encoding PR2, a β-1,3-endoglucanase; PR3, a class I basic chitinase CHN50; PR4, an endochitinase; and PR5, an osmotin (van Loon et al., 2006). In addition, diverse genes known to be preferentially expressed during plant defence were induced, such as Hin1 (Gopalan et al., 1996), SAR8.2A (Alexander et al., 1992), HSR201 (Czernic et al., 1996), and HMGR2 (Genschik et al., 1992). In accordance with this, increased expression of pathogenesis-related and SA-responsive genes upon overexpression of ATL genes has recently been reported (Serrano and Guzman, 2004; Hondo et al., 2007; Liu et al., 2008b). Another key element in the signalling cascade leading to HR downstream of R-gene-mediated pathogen recognition, EDS1, has been found to be up-regulated in the PtaRHE1 overexpressors. EDS1 encodes a lipase-like protein required for disease resistance mediated by the tobacco N protein, a TIR-NBS-LRR protein (Peart et al., 2002). Two genes coding for WRKY transcription factors were induced in the PtaRHE1 overexpressors, TIZZ (a homologue of AtWRKY40) and NtWRKY12 (a homologue of AtWRKY51). WRKY proteins are a family of transcription factors that are strongly and rapidly up-regulated in response to wounding, pathogen infection, or abiotic stresses in numerous plant species (Ülker and Somssich, 2004). Moreover, WRKY proteins have been associated with the regulation of developmental processes such as trichome and seed development (Eulgem and Somssich, 2007) as well as with leaf senescence (Balazadeh et al., 2008). WRKY factors, which ensure proper cellular responses to internal and external signals, bind to particular cis-elements found in various gene promoters and called W-boxes (Eulgem et al., 2000). W-boxes are considered to be major cis-acting elements responsible for the pathogen inducibility of many plant genes and have been found in the promoters of various wound- and pathogen-responsive genes, including several PR genes (Rushton and Somssich, 1998; Yang et al., 1999; Yu et al., 2001). Two binding sites for NtWRKY12 have been identified in the PR-1a promoter, and NtWRKY12 has been shown to activate PR-1a::GUS expression in A. thaliana protoplasts, providing evidence that NtWRKY12 is a transcriptional activator of PR-1a (van Verk et al., 2008). Therefore the activation of PR-1a may be due to the overexpression of PtaRHE1 either directly or via the induction of NtWRKY12. Interestingly, the isolated PtaRHE1 promoter contains eight putative W-boxes (Supplementary Fig. S4 at JXB online), suggesting that this gene could itself be regulated by particular WRKY factor(s). The identification of the poplar WRKY(s) binding to the W-boxes in pPtaRHE1 would indicate whether PtaRHE1 is a target for a particular WRKY and provide information on the biological function of PtaRHE1. Induction of WRKY factors and defence genes was also observed during the activation of a tobacco mitogen-activated protein kinase (MAPK) cascade (Kim and Zhang, 2004). Besides, several MAPKs, including SA-induced protein kinase (SIPK), wound-induced protein kinase (WIPK), and their upstream kinase NtMEK2, have been shown to be involved in the perception of pathogens and pathogen-derived elicitors (Zhang et al., 2000; Jin et al., 2003; del Pozo et al., 2004). In the PtaRHE1-overexpressing plants, WIPK was >2-fold induced, but the expression of NtMEK2 and SIPK was not significantly altered (Table 1). These data suggest that PtaRHE1 possibly acts upstream of WIPK in the activation of WRKYs, or in an independent pathway to trigger plant defence.
Members of the ATL gene family have already been proposed to be involved in the defence response (Craig et al., 2009). For instance, the A. thaliana ATL2 is induced following treatment by Cel, chitin, chitooctaose, and flagellin (Salinas-Mondragón et al., 1999; Navarro et al., 2004; Libault et al., 2007), the rice EL5 is up-regulated after treatment with N-acetylchitoheptaose (Takai et al., 2001), while the expression of the tobacco ACRE132 is triggered during Avr9- and Cf-9-mediated defence responses (Durrant et al., 2000). Histochemical GUS staining revealed that PtaRHE1 is induced by pathogens, SA, and Cel, suggesting that this gene might be part of the genetic network activated during plant responses to pathogens and elicitors (Fig. 4). These results are in accordance with the occurrence of W- and GT-boxes in the pPtaRHE1 sequence (see the legend of Supplementary Fig. S4 at JXB online). In addition, this promoter is activated by ABA, possibly due to the occurrence of ABREs in pPtaRHE1. Similar results have been observed in poplar leaves where PtaRHE1 was found to be clearly induced by Cel and ABA, as shown by RT-PCR analysis (Supplementary Fig. S5), indicating a comparable response of this gene to these stress conditions in both poplar and tobacco.
The GUS histochemical analysis also revealed that pPtaRHE1 is expressed in the connective tissue of anthers and in the stigma (Fig. 5). Recently, an analysis of the N. tabacum stigma/style transcriptome revealed that highly expressed genes in these tissues are associated with defence mechanisms or pollen–pistil interactions (Quiapim et al., 2009). Connective tissue has been shown to undergo PCD during development and dehiscing of the anther (Senatore et al., 2009). The spatio-temporal expression pattern of the PtaRHE1 promoter showed that PtaRHE1 is developmentally regulated, being high in young leaves and roots (Fig. 5A–D). At later stages of development and in tissues undergoing the secondary growth phase, GUS staining was localized in ray parenchyma cells (Fig. 5I). This observation is in agreement with the previously reported expression of PtaRHE1 in poplar stem, as analysed by in situ RT-PCR (van Raemdonck et al., 2005). Likewise, in zinnia, the RING-encoding gene ZeRH2.1 was shown to be expressed within vascular bundles of the mature stem in xylem parenchyma cells and in the phloem (Dahiya et al., 2005). Therefore, these authors suggested a role for ZeRH2.1 during active transport. A role for PtaRHE1 in transport could also be proposed since ray parenchyma cells are involved in the transport of water and nutrients as well as of signalling molecules between the phloem and the xylem.
Characterizing the exact function(s) of PtaRHE1 requires the identification of PtaRHE1’s target(s). Nevertheless, the pleiotropic effects observed in tobacco overexpressors suggest that PtaRHE1 targets (a) protein(s) involved in signalling cascades/pathways regulating important developmental processes and the interaction of plants with their biotic and abiotic environments. The increased expression of pathogenesis-related genes, the responsiveness of the promoter PtaRHE1 to pathogens and elicitors, as well as the HR-like phenotype induced in transgenic tobaccos suggest that PtaRHE1 might target (a) protein(s) that is/are linked to defence mechanisms but that might also regulate developmental processes at a point where both genetic networks intersect each other.
Supplementary data
Additional supporting information may be found at JXB online.
Figure S1. Alignment of the PtaRHE1 amino acid sequence with the Arabidopsis ATL2 amino acid sequence (At3g16720).
Figure S2. Sequences of full-length PtaRHE1 and PtaRHE1-Ct (where the TM domain and the basic domains were deleted).
Figure S3. RT-PCR expression analysis of PtaRHE1 in three individual plants of the WT, RLR1-1-1, and RLR1-5-7.
Figure S4. Nucleotide sequence of the 5′-flanking promoter region and putative cis-acting elements of the PtaRHE1 promoter.
Figure S5. RT-PCR expression analysis of PtaRHE1 in poplar leaves treated during 8 h with Cel (100 μg ml−1) and ABA (150 μM). 18S was used as a loading control.
Table S1. Primer sequences and amplicon size for RT-PCR and RT-qPCR. The gene classification was based on literature data.
Supplementary Material
Acknowledgments
M.B. is a Senior Research Associate of the FRS-FNRS. O.M.V. is a post-doctoral Researcher of the FRS-FNRS. This work was funded by the ‘Fonds de la Recherche Fondamentale Collective’ (no. 2.4.574.06.F).
References
- Aguilar-Henonin L, Bravo J, Guzmán P. Genetic interactions of a putative Arabidopsis thaliana ubiquitin-ligase with components of the Saccharomyces cerevisiae ubiquitination machinery. Current Genetics. 2006;50:257–268. doi: 10.1007/s00294-006-0093-y. [DOI] [PubMed] [Google Scholar]
- Alexander D, Stinson J, Pear J, Glascock C, Ward E, Goodman RM, Ryals J. A new multigene family inducible by tobacco mosaic virus or salicylic acid in tobacco. Molecular Plant-Microbe Interactions. 1992;5:513–515. doi: 10.1094/mpmi-5-513. [DOI] [PubMed] [Google Scholar]
- Bachmair A, Becker F, Materson RV, Schell J. Perturbation of the ubiquitin system causes leaf curling, vascular tissue alterations and necrotic lesions in a higher plant. EMBO Journal. 1990;9:4543–4549. doi: 10.1002/j.1460-2075.1990.tb07906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends in Plant Science. 2001;6:463–470. doi: 10.1016/s1360-1385(01)02080-5. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biology. 2008;10:63–75. doi: 10.1111/j.1438-8677.2008.00088.x. [DOI] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. The Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffey N, Cholewa E, Regan S, Sundberg B. Secondary xylem development in Arabidopsis: a model for wood formation. Physiologia Plantarum. 2002;114:594–600. doi: 10.1034/j.1399-3054.2002.1140413.x. [DOI] [PubMed] [Google Scholar]
- Craig A, Ewan R, Mesmar J, Gudipati V, Sadanandom A. E3 ubiquitin ligases and plant innate immunity. Journal of Experimental Botany. 2009;60:1123–1132. doi: 10.1093/jxb/erp059. [DOI] [PubMed] [Google Scholar]
- Czernic P, Huang HC, Marco Y. Characterization of hsr201 and hsr515, two tobacco genes preferentially expressed during the hypersensitive reaction provoked by phytopathogenic bacteria. Plant Molecular Biology. 1996;31:255–265. doi: 10.1007/BF00021788. [DOI] [PubMed] [Google Scholar]
- Dahiya P, Milioni D, Wells B, Stacey N, Roberts K, McCann M. A RING domain gene is expressed in different cell types of leaf trace, stem, and juvenile bundles in the stem vascular system of zinnia. Plant Physiology. 2005;138:1383–1395. doi: 10.1104/pp.104.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Reynaerts A, Höfte H, Hernalsteens J-P, Leemans J, Van Montagu M. Vectors for cloning in plant cells. Methods in Enzymology. 1987;153:277–292. [Google Scholar]
- del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO Journal. 2004;23:3072–3082. doi: 10.1038/sj.emboj.7600283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Current Biology. 2006;16:272–279. doi: 10.1016/j.cub.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. The Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich E. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in Plant Science. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. The Plant Cell. 2007;19:417–432. doi: 10.1105/tpc.106.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Marbach J, Durr A, Fleck J, Jamet E. Isolation and characterization of a cDNA encoding a 3-hydroxy-3-methylglutaryl coenzyme A reductase from Nicotiana sylvestris. Plant Molecular Biology. 1992;20:337–341. doi: 10.1007/BF00014504. [DOI] [PubMed] [Google Scholar]
- Gopalan S, Wei W, He SY. hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. The Plant Journal. 1996;10:591–600. doi: 10.1046/j.1365-313x.1996.10040591.x. [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic Y. Ubiquitylation and cell signaling. EMBO Journal. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. cdc2a expression in A. thaliana is linked with competence for cell division. The Plant Cell. 1993;5:1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondo D, Hase S, Kanayama Y, Yoshikawa N, Takenaka S, Takahashi H. The LeATL6-associated ubiquitin/proteasome system may contribute to fungal elicitor-activated defense response via the jasmonic acid-dependent signaling pathway in tomato. Molecular Plant-Microbe Interactions. 2007;20:72–81. doi: 10.1094/MPMI-20-0072. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Jensen KL, Jespersen HM, Skriver K. Widespread occurrence of a highly conserved RING-H2 zinc finger motif in the model plant Arabidopsis thaliana. FEBS Letters. 1998;436:283–287. doi: 10.1016/s0014-5793(98)01143-0. [DOI] [PubMed] [Google Scholar]
- Jin H, Liu Y, Yang K-Y, Kim CY, Baker B, Zhang S. Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. The Plant Journal. 2003;33:719–731. doi: 10.1046/j.1365-313x.2003.01664.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Karlowski WM, Hirsch AM. The over-expression of an alfalfa RING-H2 gene induces pleiotropic effects on plant growth and development. Plant Molecular Biology. 2003;52:121–133. doi: 10.1023/a:1023916701669. [DOI] [PubMed] [Google Scholar]
- Katoh S, Hong C, Tsunoda Y, Murata K, Takai R, Minami E, Yamazaki T, Katoh E. High precision NMR structure and function of the RING-H2 finger domain of EL5, a rice protein whose expression is increased upon exposure to pathogen-derived oligosaccharides. Journal of Biological Chemistry. 2003;278:15341–15348. doi: 10.1074/jbc.M210531200. [DOI] [PubMed] [Google Scholar]
- Katoh S, Tsunoda Y, Murata K, Minami E, Katoh E. Active site residues and amino acid specificity of the ubiquitin carrier protein-binding RING-H2 finger domain. Journal of Biological Chemistry. 2005;280:41015–41024. doi: 10.1074/jbc.M411127200. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Nam J, Boyes DC, Holt BF, III, Hubert DA, Wiig A, Dangl JL. A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. The Plant Journal. 2005;44:258–270. doi: 10.1111/j.1365-313X.2005.02525.x. [DOI] [PubMed] [Google Scholar]
- Kim CY, Zhang S. Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. The Plant Journal. 2004;38:142–151. doi: 10.1111/j.1365-313X.2004.02033.x. [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn J-W, Jin U-H, Choi D, Paek K-H, Pai H-S. Activation of the programmed cell death pathway by inhibition of proteasome function in plants. Journal of Biological Chemistry. 2003;278:19406–19415. doi: 10.1074/jbc.M210539200. [DOI] [PubMed] [Google Scholar]
- Kim M, Lee S, Park K, Jeong E-J, Ryu C-M, Choi D, Pai H-S. Comparative microarray analysis of programmed cell death induced by proteasome malfunction and hypersensitive response in plants. Biochemical and Biophysical Research Communications. 2006;342:514–521. doi: 10.1016/j.bbrc.2006.01.176. [DOI] [PubMed] [Google Scholar]
- Ko J-H, Yang SH, Han K-H. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. The Plant Journal. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- Koiwai H, Tagiri A, Katoh S, Katoh E, Ichikawa H, Minami E, Nishizawa Y. RING-H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. The Plant Journal. 2007;51:92–104. doi: 10.1111/j.1365-313X.2007.03120.x. [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau O-S, Deng X-W, Callis J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiology. 2005;139:1597–1611. doi: 10.1104/pp.105.067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HXL, Gong Z, Ishitani M, Stevenson B, Zhu J-K. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes & Development. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin ligase genes responding to chitin, a plant defense elicitor. Molecular Plant-Microbe Interactions. 2007;20:900–911. doi: 10.1094/MPMI-20-8-0900. [DOI] [PubMed] [Google Scholar]
- Lin S-S, Martin R, Mongrand S, Vandenabeele S, Chen K-C, Jang I-C, Chua N-H. RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. The Plant Journal. 2008;56:550–561. doi: 10.1111/j.1365-313X.2008.03625.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Qin G, et al. Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. The Plant Cell. 2008a;20:1538–1554. doi: 10.1105/tpc.108.059741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Yang Y, Li G, Yang Y, Wang X, Basnayake BMVS, Li D, Song F. Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Molecular Biology. 2008b;68:17–30. doi: 10.1007/s11103-008-9349-x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCTmethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martínez-García M, Garcidueñas-Piña C, Guzmán P. Gene isolation in Arabidopsis thaliana by conditional overexpression of cDNAs toxic to Saccharomyces cerevisiae: identification of a novel early response zinc-finger gene. Molecular and General Genetics. 1996;252:587–596. doi: 10.1007/BF02172405. [DOI] [PubMed] [Google Scholar]
- Molnár G, Bancoş S, Nagy F, Szekeres M. Characterization of BRH1, a brassinosteroid-responsive RING-H2 gene from Arabidopsis thaliana. Planta. 2002;215:127–133. doi: 10.1007/s00425-001-0723-z. [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. The ubiquitin–proteasome pathway and plant development. The Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JDG. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiology. 2004;135:1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolay R, Wiederkehr T, Rist W, Kramer G, Mayer MP, Bukau B. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. Journal of Biological Chemistry. 2004;279:2673–2678. doi: 10.1074/jbc.M311112200. [DOI] [PubMed] [Google Scholar]
- Nodzon LA, Xu W-H, Wang Y, Pi L-Y, Chakrabarty PK, Song WY. The ubiquitin ligase XBAT32 regulates lateral root development in Arabidopsis. The Plant Journal. 2004;40:996–1006. doi: 10.1111/j.1365-313X.2004.02266.x. [DOI] [PubMed] [Google Scholar]
- Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC. An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. The Plant Journal. 2002;29:569–579. doi: 10.1046/j.1365-313x.2002.029005569.x. [DOI] [PubMed] [Google Scholar]
- Quiapim AC, Brito MS, Bernardes LAS, daSilva I, Malavazi I, DePaoli HC, Molfetta-Machado JB, Giuliatti S, Goldman GH, Goldman MHS. Analysis of the Nicotiana tabacum stigma/style transcriptome reveals gene expression differences between wet and dry stigma species. Plant Physiology. 2009;149:1211–1230. doi: 10.1104/pp.108.131573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE. Transcriptional control of plant genes responsive to pathogens. Current Opinion in Plant Biology. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- Salinas-Mondragón RE, Garcidueñas-Piña C, Guzmán P. Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Molecular Biology. 1999;40:579–590. doi: 10.1023/a:1006267201855. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Borden KLB, Boddy MN, Freemont PS. Does this have a familiar RING? Trends in Biochemical Science. 1996;21:208–214. [PubMed] [Google Scholar]
- Schwechheimer C, Willige BC, Zourelidou M, Dohmann EMN. Examining protein stability and its relevance for plant growth and development. Methods in Molecular Biology. 2009;479:147–171. doi: 10.1007/978-1-59745-289-2_10. [DOI] [PubMed] [Google Scholar]
- Senatore A, Trobacher CP, Greenwood JS. Ricinosomes predict programmed cell death leading to anther dehiscence in tomato. Plant Physiology. 2009;149:775–790. doi: 10.1104/pp.108.132720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Guzmán P. Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics. 2004;167:919–929. doi: 10.1534/genetics.104.028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Parra S, Alcaraz LD, Guzmán P. The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. Journal of Molecular Evolution. 2006;62:434–445. doi: 10.1007/s00239-005-0038-y. [DOI] [PubMed] [Google Scholar]
- Sonoda Y, Yao S-G, Sako K, Sato T, Kato W, Ohto M-A, Ichikawa T, Matsui M, Yamaguchi J, Ikeda A. SHA1, a novel RING finger protein, functions in shoot apical meristem maintenance in Arabidopsis. The Plant Journal. 2007;50:586–596. doi: 10.1111/j.1365-313X.2007.03062.x. [DOI] [PubMed] [Google Scholar]
- Stone SL, Callis J. Ubiquitin ligases mediate growth and development by promoting protein death. Current Opinion in Plant Biology. 2007;10:624–632. doi: 10.1016/j.pbi.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiology. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development is involved in abscisic acid signaling. The Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Kim B-H, Lyssenko NN, Xu X, Johnson CH, von Arnim AG. The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proceedings of the National Academy of Sciences, USA. 2004;101:6798–6802. doi: 10.1073/pnas.0307964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai R, Hasegawa K, Kaku H, Shibuya N, Minami E. Isolation and analysis of expression mechanisms of a rice gene, EL5, which shows structural similarity to ATL family from Arabidopsis, in response to N-acetylchitooligosaccharide elicitor. Plant Science. 2001;160:577–583. doi: 10.1016/s0168-9452(00)00390-3. [DOI] [PubMed] [Google Scholar]
- Takai R, Matsuda N, Nakano A, Hasegawa K, Akimoto C, Shibuya N, Minami E. EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. The Plant Journal. 2002;30:447–455. doi: 10.1046/j.1365-313x.2002.01299.x. [DOI] [PubMed] [Google Scholar]
- Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Current Opinion in Plant Biology. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Vandeputte O, Oukouomi Lowe Y, Burssens S, van Raemdonck D, Hutin D, Boniver D, Geelen D, El Jaziri M, Baucher M. The tobacco Ntann12 gene, encoding an annexin, is induced upon Rhodococcus fascians infection and during leafy gall development. Molecular Plant Pathology. 2007;8:185–194. doi: 10.1111/j.1364-3703.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- van Raemdonck D, Pesquet E, Cloquet S, Beeckman H, Boerjan W, Goffner D, El Jaziri M, Baucher M. Molecular changes associated with the setting up of secondary growth in aspen. Journal of Experimental Botany. 2005;56:2211–2227. doi: 10.1093/jxb/eri221. [DOI] [PubMed] [Google Scholar]
- van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJM. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiology. 2008;146:1983–1995. doi: 10.1104/pp.107.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viestra RD. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends in Plant Science. 2003;8:135–142. doi: 10.1016/S1360-1385(03)00014-1. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W. Light inactivation of arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux J-P, Ryals JA. Coordinate gene activity in response to agents that induce systemic acquired resistance. The Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirdnam C, Motoyama A, Arn-Bouldoires E, van Eeden S, Iglesias A, Meins F., Jr Altered expression of an ankyrin-repeat protein results in leaf abnormalities, necrotic lesions, and the elaboration of a systemic signal. Plant Molecular Biology. 2004;56:717–730. doi: 10.1007/s11103-004-4679-9. [DOI] [PubMed] [Google Scholar]
- Xie Q, Guo H-S, Dallman G, Fang S, Weissman AM, Chua N-H. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 2002;419:167–170. doi: 10.1038/nature00998. [DOI] [PubMed] [Google Scholar]
- Xu R, Li QQ. A RING-H2 zinc-finger protein gene RIE1 is essential for seed development in Arabidopsis. Plant Molecular Biology. 2003;53:37–50. doi: 10.1023/b:plan.0000009256.01620.a6. [DOI] [PubMed] [Google Scholar]
- Yaeno T, Iba K. BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiology. 2008;148:1032–1041. doi: 10.1104/pp.108.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Chen C, Wang Z, Fan B, Chen Z. A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. The Plant Journal. 1999;18:141–149. [Google Scholar]
- Yu D, Chen C, Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. The Plant Cell. 2001;13:1527–1539. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu Y, Klessig DF. Multiple levels of tobacco WIPK activation during the induction of cell death by fungal elicitins. The Plant Journal. 2000;23:339–347. doi: 10.1046/j.1365-313x.2000.00780.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua N-H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes & Development. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. The Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng LR, Vega-Sánchez ME, Zhu T, Wang G-L. Ubiquitination-mediated protein degradation and modification: an emerging theme in plant–microbe interactions. Cell Research. 2006;16:413–426. doi: 10.1038/sj.cr.7310053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.