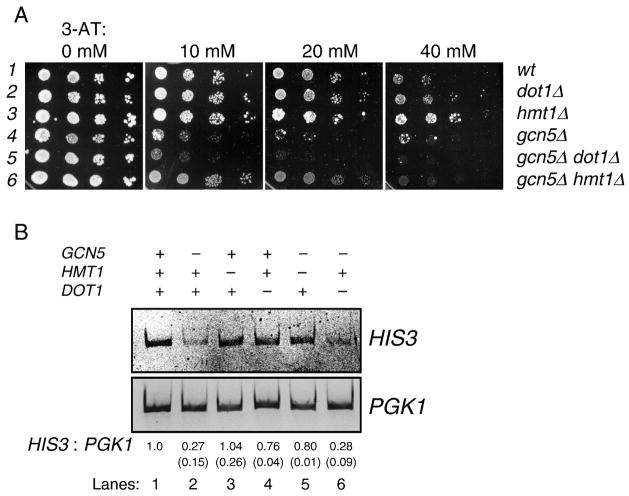
Figure 3. Gcn5p antagonizes Hmt1p for HIS3 activation.
A. Cellular growth in the presence of varying concentrations of 3-AT. The concentrations of 3-AT in synthetic complete medium minus histidine (SC –His) are listed above each panel, and the relevant genotypes are on the right. Log-phase cells in rich medium were serially diluted and spotted to the 3-AT plates. Cells were grown at 30°C for three to five days. B. HIS3 transcriptional defect caused by deleting GCN5 can be rescued by HMT1 knockout. Semi-quantitative reverse transcription-PCR (RT- PCR) was used to compare the relative amount of HIS3 transcripts, using PGK1 as the control. Log-phase yeast cells were transferred to minimal medium containing 3-AT to induce HIS3 transcription. The ratio of HIS3/PGK1 of each sample was quantified and normalized to the wildtype (lane 1) strain. Quantification data were from two independent experiments and the errors are shown in parentheses under the mean.