Abstract
Little is known about the role of protein dynamics in directing protein unfolding along a specific pathway and about the role played by chemical denaturants in modulating the dynamics and the initiation of unfolding. In this study, deuterium-hydrogen exchange (HX) detected by electrospray ionization mass spectrometry (ESI-MS) was used to study the unfolding of the SH3 domain of the PI3 kinase. Unfolding on the principal unfolding pathway occurs in 2 steps, both in the absence and in the presence of 1.8 M guanidine hydrochloride (GdnHCl). In both cases, the first step leads to the formation of an intermediate, IN, with 5 fewer protected amide hydrogen sites than in N. In the second step, IN loses the structure protecting the remaining 14 amide hydrogen sites from HX as it unfolds completely. ESI-MS analysis of fragments of the protein created by proteolytic digestion, after completion of the HX reaction, shows that IN has lost protection against HX in the same segments of native structure during unfolding in the absence and presence of 1.8 M GdnHCl. Hence, GdnHCl does not appear to play a direct active role in the initiation of unfolding. However, at higher GdnHCl concentrations, a second unfolding pathway is shown to compete effectively with the N ↔ IN ↔ U pathway. In this way, the denaturant modulates the energy landscape of unfolding.
Keywords: hydrogen exchange, mass spectrometry, protein unfolding, partially unfolded conformation, native-state exchange
Folded proteins possess dynamic structures, and the lowest energy conformation of a folded protein exists at equilibrium with many high energy conformational substates, which are Boltzmann-distributed in the folded protein ensemble (1–4). Fluctuations in protein structure appear not to be random. They may be directed toward enhancing function (3–7), but their role in preferentially directing protein folding and unfolding reactions on to specific pathways is poorly understood (8). Even less is known about how the thermal fluctuations that lead to protein unfolding are perturbed by denaturants such as guanidine hydrochloride (GdnHCl) and urea. Protein denaturants may act indirectly by disrupting the structure of water, thereby making hydrophobic groups more readily solvated (9, 10), or directly by interacting more strongly than water with the protein backbone and side chains (11–13). To understand how denaturants act, it is necessary to determine whether the mechanism of unfolding of a protein, as well as the thermal fluctuations that enable unfolding, are the same in the absence of a chemical denaturant and in the presence of a high concentration of the denaturant.
Structural characterization of protein unfolding in the presence, as well as absence, of denaturant becomes possible when the hydrogen exchange (HX) experiment is carried out in conjunction with NMR spectroscopy or MS. HX-NMR experiments have been carried out on many different proteins under conditions where HX is rate-limited by the intrinsic exchange rate constant (kint) of the fully solvent-exposed amide hydrogens (the EX2 limit), and they have enabled the structural characterization of partially unfolded forms (PUFs) in equilibrium with native protein (14–17). But such studies cannot reveal the temporal order in which the PUFs form from the native protein. The temporal order is revealed when HX experiments are carried out in conditions where exchange is rate-limited by the rate constants of conformational change and not by kint (the EX1 limit) (18–20). But so far, HX experiments, carried out both in the absence and in the presence of a high concentration of denaturant on an appropriately chosen protein, have been unable to demonstrate directly that the same partially unfolded conformation is populated initially under both conditions because of the difficulty in ensuring that HX occurs in the EX1 limit in the absence of both denaturant and high pH.
SH3 domains have long been exploited as archetypal “2-state” folders in protein folding studies, particularly for investigating transition states (21, 22), but recent investigations have revealed that folding intermediates are populated both before (23, 24) and after (25) the rate-limiting step in folding. HX-MS measurements have shown that SH3 domains sample partially unfolded conformations in the native state ensemble and even the fully unfolded conformation, in native-like conditions (15, 26, 27). The SH3 domain of the PI3 kinase is the slowest folding member of the SH3 domain family (28), suggesting the possibility that HX into the protein might occur in the EX1 limit even in the absence of denaturant at pH 7.2, 25 °C (29, 30). Moreover, a kinetic intermediate populated on the direct pathway of unfolding of this SH3 domain (25), can potentially serve as the signpost on the unfolding pathway for asking whether unfolding in zero and in high denaturant concentrations occurs by the same pathway (31).
In this report, it is shown that native state HX into the SH3 domain of the PI3 kinase indeed occurs within the EX1 limit. In the absence of denaturant, unfolding occurs in 2 principal steps. In the first step, 5 of the 19 amide hydrogen sites protected against HX in the folded protein, lose their protection, and an intermediate, IN, with 14 protected amide hydrogen sites is populated. In the second step, all amide hydrogen sites become deprotected during transient formation of U. In high denaturant concentration, the principal unfolding reaction also occurs in 2 steps: the intermediate IN, with 14 protected amide hydrogens, is now populated within a few seconds of unfolding. Proteolytic analysis of the locations of the protected amide hydrogen sites in IN formed in the absence and presence of denaturant suggests that this initial unfolding intermediate has the same structure in both unfolding conditions.
Results and Discussion
In this study, HX studies were carried out at pH 7.2 and 25 °C, by diluting the deuterated protein 15-fold into labeling buffer, either containing 0, 0.5, or 1 M GdnHCl in which the protein remains native (N) at equilibrium or containing 1.8 M GdnHCl in which >95% of the protein molecules are unfolded (U) at equilibrium. After increasing durations of labeling, exchange was quenched at pH 2.6, and the samples were processed for ESI-MS. An apparently unimodal spectrum (isotope distribution) is seen for N, which is centered at a mass indicating that N has 19 ± 1 amide hydrogen sites that are protected from HX. All other amide hydrogen sites undergo HX because of weak protection under the labeling and sample work-up conditions used in this study.
HX During Unfolding in Strongly Destabilizing Conditions.
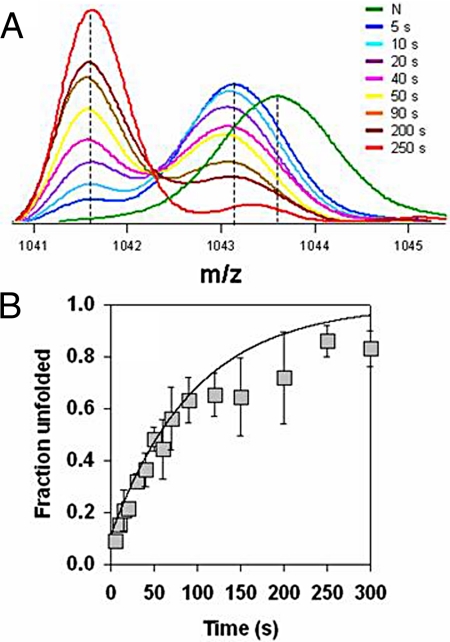
Fig. 1A shows mass spectra collected at different times of HX/unfolding in 1.8 M GdnHCl. At the earliest time (5 s) of HX/unfolding itself, the unimodal mass profile characteristic of the 19 deuteriums-containing N from which unfolding began has changed significantly. It has become bimodal, with approximately 15% of the mass distribution in a lower mass peak and approximately 85% in a higher mass peak. Importantly, the higher mass peak is not that of N with 19 protected deuteriums, but of a native-like intermediate conformation, henceforth referred to as IN, with only 14 protected deuteriums consequent to 5 deuteriums having exchanged out. Hence, 85% of the protein molecules have partially unfolded to IN at 5 s of unfolding in 1.8 M GdnHCl. The lower mass peak indicates that at this time about 15% of the protein molecules have unfolded to a conformation that is fully protonated. The mass spectrum is bimodal because the intermediate appears to unfold to a fully exchange-competent conformation in one slow step and because the intermediate and unfolded conformations differ in mass by 14 Da, resulting in their mass distributions being well separated (30). With time of unfolding, the mass spectrum remains bimodal, and the lower mass peak gains in intensity at the expense of the higher mass peak. At each time of unfolding, the mass profile fits well to the sum of 2 Gaussian distributions. Finally, the 2 peaks collapse into a single lower mass peak, as all molecules become completely protonated at all of the 19 amide hydrogen sites that are protected in the N, except 2 deuteriums that remain bound due to 7% residual deuterium present during labeling.
Fig. 1.
Unfolding kinetics of the SH3 domain in 1.8 M GdnHCl, pH 7.2 monitored by HX-MS. Exchange was allowed to proceed in 1.8 M GdnHCl for different times. (A) shows representative spectra obtained at different times of unfolding. The mass spectrum shown in green corresponds to the native-state having 19 ± 1 deuteriums protected and was obtained by quenching the exchange after 5 s of labeling in the absence of GdnHCl. The mass spectrum for the unfolded-state is shown in red. All of the mass spectra correspond to the + 9 charge state. (B) shows the increase in the fraction of completely protonated molecules with the time of unfolding. The error bars represent the standard deviations from 3 separate experiments. The solid line through the data represents a kinetic simulation to the 2-pathway mechanism using values for the rate constants as shown (Fig. S1).
Fig. 1B shows that the change in the fractional population of completely protonated molecules occurs with biphasic kinetics: an approximately 15% burst phase change is followed by a slow apparently exponential phase. For unfolding in 1.8 M GdnHCl, it is known from optical studies that unfolding occurs via an N ↔ IU ↔ U mechanism and that the on-pathway intermediate IU is populated to an extent of 15% at 10 ms of unfolding (25). Hence, the slow exponential phase appears to correspond to the formation of U, and the burst phase corresponds to the accumulation of IU, which, like U, has no protected amide hydrogen sites. This is not surprising because previous studies have shown that although IU is N-like in its far-UV circular dichroism properties, it nevertheless is U-like in its fluorescence properties and it has considerably more solvent-exposed surface area than N (25).
The native-like intermediate IN had not been detected in previous optical studies (25) of the unfolding because its fluorescence and circular dichroism properties are the same as that of N. Hence, it is necessary to position IN correctly with respect to the N ↔ IU ↔ U pathway. Kinetic simulations were carried out to decide whether (i) IN is on-pathway in an N ↔ IN ↔ IU ↔ U mechanism, (ii) off-pathway in an IN ↔ N ↔ IU ↔ U mechanism, (iii) off-pathway but forming from IU in a branched mechanism (supporting information (SI) Text), or (iv) whether it defines a second unfolding pathway N ↔ IN ↔ U which competes with the N ↔ IU ↔ U pathway in a 2-pathway mechanism (SI Text). These simulations indicate that the 2-pathway mechanism (SI Text and Fig. S1) is sufficient to explain not only the data in Fig. 1 but also the previously obtained optical data on unfolding (25). Importantly they rule out the alternative mechanisms (SI Text).
Native-State HX-MS Also Reveals Two Stages of Unfolding.
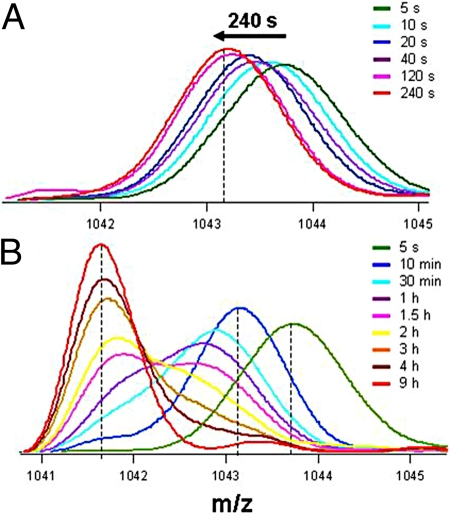
In native-like conditions, the frequency of the unfolding events leading to the transient formation of IN, IU, and U will be very low. Nevertheless, the kinetics of unfolding can be measured by appropriate native-state HX measurements, here carried out in 0 M (Fig. 2 A and B), 0.5 M (Fig. S2 a and b), and 1.0 M (Fig. S2 c and d) GdnHCl at pH 7.2, 25 °C. In all 3 conditions, the mass spectra at different times of HX/unfolding are bimodal (see next page). But during the time taken for the first 5 deuteriums to exchange out, which decreases from 240 s in 0 M GdnHCl to 20 s in 1 M GdnHCl (Figs. 2 and S2), the relative intensity of the lower mass peak is very low (<5% of the molecules have become fully protonated); hence, the mass profile is essentially unimodal during this time. As the mass profile migrates during the time the first 5 deuteriums take to exchange out, its width does not change.
Fig. 2.
Kinetics of native-state exchange of the SH3 domain at pH 7.2. Exchange was allowed to proceed for different times in 0 M GdnHCl. The native-state spectra shown in green in (A and B) were obtained as described in the legend to Fig. 1. The mass spectrum shown by the red line in (B) corresponds to the completely unfolded state, which retains 2 deuteriums due to the presence of residual (7%) D2O during labeling. (A) shows the unimodal mass spectra for exchange of the first 5 deuteriums, at different times of labeling. The mass spectra shift gradually to the m/z value of IN during the time indicated above the arrow. (B) shows representative mass profiles for the exchange of the remaining 14 protected deuteriums, at different times of labeling. The mass spectra in both the panels correspond to the + 9 charge state.
In native-like conditions, unlike in 1.8 M GdnHCl (Fig. 1), the exchange-out of the first 5 deuteriums upon transient unfolding to IN is slow enough to be resolved temporally. Two observations concerning the time taken for the 5 deuteriums to exchange out, which decreases significantly with an increase in GdnHCl concentration (Figs. 2 and S2), suggest that HX into IN occurs by the EX1 mechanism. Firstly, this observed time is much larger than the time constant for intrinsic exchange, kint−1. Hence, the rate constant of the IN → N transition, which would necessarily be faster than that of the N → IN transition in native conditions, must be ≪kint. This fulfills a necessary condition for HX to occur in the EX1 limit. Secondly, this observed time is the same whether HX is carried out at pH 8.2 or at pH 7.2 (Fig. S3). If HX were occurring in the EX2 limit, the time taken for the 5 deuteriums to exchange out would be substantially shorter at the higher pH because kint is 10-fold higher at pH 8.2 than at pH 7.2 (SI Text). On the other hand, the observed pH independence is expected when HX into IN occurs in the EX1 limit, because the stability and unfolding kinetics of the protein are identical at both pH values (Fig. S4).
Hence, it appears that HX into IN must occur in the EX1 limit even though essentially unimodal mass spectra are seen. For the turkey ovomucoid third domain, unimodal mass spectra were also observed when HX occurs in the EX1 limit (18, 32). In this study, the shift in the apparently unimodal mass profile during the exchange-out of the first 5 deuteriums can be explained if transient unfolding to IN occurs in more than 1 step, with 1–2 amide hydrogen sites losing protection in each step. Fig. S5 shows that the mass spectrum at any intermediate time during the migration of the apparently unimodal mass peak in zero denaturant is not describable as the weighted sum of the mass spectra of N, which has 19 protected amide deuteriums, and IN, which has 14 protected amide deuteriums (see SI Text). Similarly, the initial stages of exchange out in 0.5 and 1 M GdnHCl at pH 7.2, and in 0 M GdnHCl at pH 8.2, are also not describable by a 2-state transition between N and IN. Exchange-out of the 5 deuteriums during the N → IN transition in more than one step is possible if the deuteriums are distributed in at least 2 distinct regions of the protein structure, which unfold either sequentially or independently of each other.
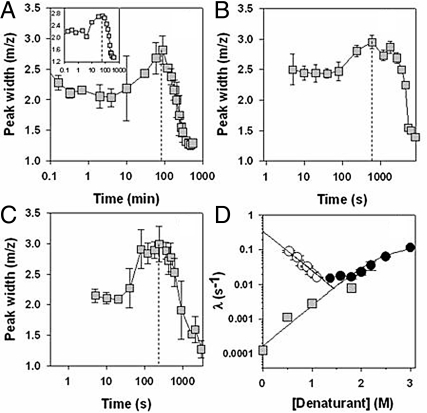
Further evolution of the mass spectra, as all molecules become completely protonated with increasing time of HX, proceeds in a more pronounced bimodal manner. The higher mass and lower mass peaks both change not only in relative intensity but also in position and width. In such cases, an estimate of the half-life (t1/2) of the observed exchange process can be obtained as the time of exchange at which the width of the mass distribution is at maximum (see Data Analysis). Fig. 3 shows how the overall peak width changes with time of exchange in 0, 0.5, and 1 M GdnHCl, at pH 7.2, and in 0 M GdnHCl, at pH 8.2. The observed rate constants (0.693/t1/2) of formation of fully protonated molecules in 0, 0.5, and 1 M GdnHCl fall reasonably well on the extrapolated dependence on GdnHCl concentration of the fluorescence-monitored rate constants for unfolding to U (Fig. 3D) at pH 7.2. Hence, the observed rate constants for complete protonation appear to be the effective rate constants of unfolding to U, indicating that HX at the 14 amide hydrogen sites that were protected in IN occurs into U. Since the observed rate constant is about the same as the rate constant of unfolding to U, HX, into U must occur in the EX1 limit, as already suggested by the bimodal nature of the mass spectra. HX into U is expected to occur in the EX1 limit because the observed rate constant of the folding of U is estimated to be 0.3 s−1 in 0 M GdnHCl, 0.09 s−1 in 0.5 M GdnHCl, and 0.025 s−1 in 1 M GdnHCl (25). These values are all much smaller than the value of kint (SI Text), which is a necessary condition for HX to occur in the EX1 limit. The EX1 nature of the HX reaction is confirmed by its pH independence: the observed rate constant of HX/unfolding in 0 M GdnHCl is the same at pH 7.2 and pH 8.2 (Figs. 2 and S3). The demonstration that HX in the EX1 limit is truly responsible for the bimodal mass spectra seen during exchange into U in native-like conditions is important because EX1 behavior can sometimes arise from uncorrelated unfolding smaller than global unfolding (30).
Fig. 3.
Peak width analysis of the mass profiles. (A, B, and C) The data for 0 M, 0.5 M, and 1.0 M GdnHCl, respectively. The peak width was determined at 15% intensity of the peak maximum. The vertical dashed lines denote the half-life of unfolding. (A, Inset) shows the half-life of unfolding when exchange was carried out in 0 M GdnHCl at pH 8.2. (D) shows the GdnHCl dependence of the observed unfolding rate constants obtained from HX-MS (▩) and when unfolding was monitored by the change in intrinsic fluorescence (●). (○) shows the observed refolding rate constants monitored by the change in intrinsic fluorescence. The fits through the observed unfolding and refolding rate constants are according to the 3-state mechanism (25). The fluorescence data are from ref. 25.
It should be noted that the observed rate constant of HX in 1.8 M GdnHCl is also predicted (within a factor of ≈2) by the fluorescence-monitored rate constant of unfolding to U. It is not surprising that HX into U occurs in the EX1 limit during unfolding in 1.8 M GdnHCl, when it does so in 0, 0.5, and 1 M GdnHCl, because U folds much more slowly in 1.8 M GdnHCl. Moreover, an iso-m/z point is observed for the mass spectra describing HX during unfolding in 1.8 M GdnHCl (Fig. 1), pointing to a 2-state unfolding reaction from IN to U. The absence of a similar iso-m/z point for mass spectra describing HX in native-like conditions (Figs. 2 and S2), suggests that in addition to transient global unfolding events (to U) many local unfolding events also occur in native-like conditions. In 1.8 M GdnHCl, these local unfolding events are not seen because the global unfolding events dominate.
In native-like conditions, the population of completely protonated molecules increases in an apparently exponential manner with time of HX, and the observed rate constants of HX/unfolding extracted from the exponential time dependences (Fig. S6) agree well (within a factor of ≈2) with those extracted from peak width analysis (Fig. 3). More importantly, there is no evidence for a weak start (lag) to the increase in the population of completely protonated molecules (Fig. S6). The kinetics of unfolding/HX (Fig. S6) could be simulated well using the 2-pathway mechanism (SI Text and Fig. S1). The simulations indicated that in native-like conditions, all N molecules use the N ↔ IN ↔ U pathway to sample IN and U. The N ↔ IU ↔ U pathway remains essentially inoperative probably because the rate constants defining it are much smaller than those defining the N ↔ IN ↔ U in native like conditions. It should be noted that unlike in the case of HX experiments in 1.8 M GdnHCl, where it was possible to demonstrate that IN is on-pathway and not off-pathway (see above), the HX experiments in zero and low denaturant concentrations by themselves do not allow such a delineation to be made. To be able to make such a delineation, it is first necessary to show that the same 5 amide hydrogen sites become deprotected during the formation of IN in both high denaturant and native-like conditions.
The Structure of IN Is Similar in Zero Denaturant and in 1.8 M GdnHCl.
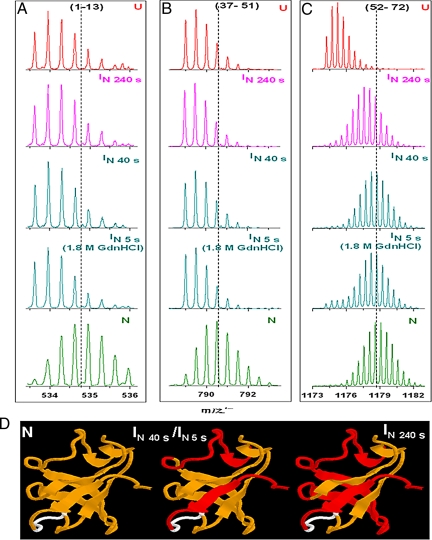
In HX-MS studies, the locations along the sequence of the amino acid residues whose amide hydrogen sites are protected, or exchanged, can be identified by fragmenting the protein by proteolysis after the exchange reaction is complete. The proteolysis is carried out under low pH conditions to minimize back exchange. Fig. S7 shows the pattern of fragments obtained by pepsin treatment of the protonated SH3 domain at pH 2.6. The 6 fragments were identified by exact mass measurement and collision induced dissociation (CID) tandem mass spectrometry (MS/MS) (Table 1) and are seen to cover 93% of the sequence. To identify the regions of the protein which are unfolded (exchanged-out) in an intermediate populated at any time during unfolding, the masses (which indicate the deuterium content) of the 6 proteolytic fragments made from a sample corresponding to the intermediate, are compared to the masses of the corresponding fragments made from N and U.
Table 1.
Deuterium levels of peptides originating from N, and from IN at 40 and 240 s of unfolding in 0 M GdnHCl and at 5 s of unfolding in 1.8 M GdnHCl
| Fragment | 1–13 | 21–29 | 24–36 | 37–51 | 52–72 | 73–83 |
|---|---|---|---|---|---|---|
| N | 1.48 ± 0.21 | 1.91 ± 0.06 | 2.46 ± 0.48 | 1.78 ± 0.01 | 7.06 ± 0.20 | 1.45 ± 0.16 |
| IN (40 s) in 0 M GdnHCl | 0.01 ± 0.21 | 1.98 ± 0.02 | 2.77 ± 0.08 | 0.21 ± 0.03 | 6.33 ± 0.09 | 0.94 ± 0.02 |
| N-IN (40 s) | 1.47 | −0.06 | −0.31 | 1.57 | 0.73 | 0.50 |
| IN (240 s) in 0 M GdnHCl | −0.19 ± 0.14 | 1.92 ± 0.04 | 2.50 ± 0.12 | −0.10 ± 0.08 | 5.2 ± 0.21 | 0.58 ± 0.07 |
| N-IN (240 s) | 1.67 | −0.003 | −0.04 | 1.89 | 1.86 | 0.87 |
| IN (5 s) in 1.8 MGdnHCl | −0.33 ± 0.11 | 1.99 ± 0.02 | 2.71 ± 0.06 | −0.03 ± 0.06 | 6.32 ± 0.19 | 0.94 ± 0.05 |
| N-IN (5 s) | 1.81 | −0.07 | −0.25 | 1.82 | 0.74 | 0.51 |
The number of deuteriums exchanged in a fragment from the sample corresponding to an intermediate are obtained by subtracting the number of deuteriums retained in that fragment from number of deuteriums protected in the corresponding fragment from native protein.
Fig. 4 shows representative mass spectra of the fragments 1–13, 37–51, and 52–72 made from deuterated N, the intermediate IN at 40 and 240 s of HX/unfolding in zero denaturant at pH 7.2, and from U. The mass distributions and average m/z values of fragments 1–13 and 37–51 from IN at both times of HX/unfolding are similar to those of U, indicating that the regions of the protein corresponding to these fragments have lost their protective structure at 40 s. On the other hand, the mass distribution and average m/z value of fragment 52–72 from IN at 40 s is N-like but becomes U-like when IN is fully formed at 240 s, indicating that the region of the protein corresponding to this fragment loses some of its protective structure between 40 and 240 s of HX/unfolding. The mass distributions and average m/z values of these fragments from IN at 5 s of unfolding in 1.8 M GdnHCl are similar to those of IN at 40 s of HX/unfolding in zero denaturant (Fig. 4), indicating that they have similar structures. Table 1 (which compares the deuterium contents of the 6 fragments from N from IN at 40 s, as well as at 240 s of HX/unfolding in zero denaturant, and from IN at 5 s of unfolding in 1.8 M GdnHCl) clearly shows that the same sequence segments of the native structure have unfolded in IN at 5 s of unfolding in 1.8 M GdnHCl and at 40 s of HX/unfolding in the absence of GdnHCl. Based on the locations of the deuteriums lost during the N → IN transition at 40 and 240 s of HX/unfolding in zero denaturant and at 5 s of unfolding in 1.8 M GdnHCl, the regions that have unfolded in IN have been mapped on to the native structure of the protein (Fig. 4D).
Fig. 4.
The regions of the SH3 domain involved in partial unfolding. (A, B, and C) show representative mass spectra of 3 peptic fragments derived from intact deuterated SH3 domain after exchange for 40 and 240 s in labeling buffer containing no denaturant and after exchange for 5 s in labeling buffer containing 1.8 M GdnHCl. Peptic fragments from native and unfolded references are also shown. The dotted lines indicate the average m/z values of peptic fragments derived from the sample corresponding to the native state. (D) shows the locations of protected deuteriums in N, IN at 40 s in zero denaturant [IN (40 s)] or at 5 s in 1.8 M GdnHCl [IN (5 s)], and IN at 240 s of exchange [IN (240 s)] in zero denaturant. Protected deuteriums are distributed throughout the folded protein. The segments colored yellow have their deuteriums protected, while segments which have exchanged all or some of their protected deuteriums are shown in red. No peptide could be identified for the region shown in white.
In several fragments, fractional levels of deuterium exchange were observed. This can possibly arise if IN is a heterogeneous ensemble of conformations, with some conformations having the fragment-segment folded and others having it unfolded.
Unfolding Begins Similarly in Zero and High Denaturant Concentration on the N ↔ IN ↔ U Pathway.
The kinetic HX studies in native-like conditions indicate N samples first IN and then U through transient sequential unfolding events that take place on the N ↔ IN ↔ U pathway. Eighty-five percent of N molecules unfold via the same pathway in 1.8 M GdnHCl. In native as well as unfolding conditions, IN is shown to have lost structure in the same segments, which in the native protein protect 5 amide hydrogen sites from HX (Table 1 and Fig. 4D). IN is shown to serve as the signpost which indicates that unfolding begins in the same regions of the molecule in the absence and presence of high denaturant. Thus, the fluctuations that lead to amide hydrogen exchange in zero and low denaturant are indeed the same as those that cause unfolding at high denaturant. This result is important because it has been suggested that this may not be so in the case of cytochrome c (33), the protein whose equilibrium unfolding has been best characterized by native-state HX.
Native-state HX studies have previously suggested that a native protein samples similar PUFs in the absence and presence of very low concentrations of denaturant (14–18). But these equilibrium studies could not directly show that these PUFs are on-pathway, and in a few cases, they have been demonstrated to be off-pathway (34). The problem of demonstrating on-pathway roles for PUFs is compounded by the results of kinetic HX studies that indicate the possibility of multiple unfolding pathways (32). The kinetic HX study on cytochrome c, which showed that the PUFs identified by equilibrium HX studies are indeed on-pathway, had to be carried out in the presence of low denaturant concentration to ensure that HX occurred in the EX1 limit (20), and hence, it could not address the question of whether the same unfolding pathway is used in the zero and high denaturant concentrations. This study demonstrates that the same initial intermediate is formed on the same (N ↔ IN ↔ U) unfolding pathway during the unfolding of the SH3 domain in the absence of denaturant and at a high denaturant concentration in which >95 of the protein molecules become fully unfolded.
Computer simulations have suggested that a denaturant can unfold a protein either by binding directly to the protein via hydrogen bonding, electrostatic, or van der Waals interactions, or by altering the solvent environment (35–38). A very recent NMR study of HX at the peptide groups of dialanine has shown that GdnHCl, unlike urea, does not hydrogen bond to the peptide group (13). The result here that the initial step in the unfolding of the SH3 domain via the N ↔ IN ↔ U pathway is similar in zero and 1.8 M GdnHCl suggests that GdnHCl does not actively attack the protein but instead acts indirectly by altering the properties of water, thereby facilitating the large intrinsic thermal fluctuations that drive unfolding on the N ↔ IN ↔ U pathway. It is also possible that the denaturant may act by stabilizing the partially unfolded, high energy conformations created by thermal fluctuations, thereby shifting the equilibrium toward unfolded conformations.
Modulation of the Unfolding Energy Landscape by Chemical Denaturant.
A molecular dynamics study of the folding of the SH3 domain had suggested that folding may occur along multiple folding pathways defined by intermediates (24). This study shows that in native-like conditions, transient unfolding occurs via the N ↔ IN ↔ U pathway but that as the GdnHCl concentration is raised, the N ↔ IU ↔ U pathway begins to effectively compete with the N ↔ IN ↔ U pathway. In very high denaturant concentration, the N ↔ IU ↔ U pathway is the predominant pathway (25). It should be noted that such switching between unfolding pathways with a change in denaturant concentration has also been observed in the case of titin (39).
Methods
Protein Purification.
The protein purification procedure has been described previously (25). The purity of the protein was checked by SDS/PAGE and ESI-MS and was found to be >98% pure. The molecular weight of the SH3 domain determined by ESI-MS is 9363.8 Da.
HX-MS Monitored Unfolding Kinetics.
For the kinetic study of unfolding in 1.8 M GdnHCl, the procedure followed was similar to that for native-state exchange except that the labeling buffer contained 1.95 M GdnHCl, so that the final GdnHCl concentration was 1.8 M. Unfolding was stopped, and exchange was quenched by diluting the GdnHCl to 0.9 M, while simultaneously lowering the pH to 2.6 by addition of an equal volume of the quench buffer.
Native-State Exchange at pH 7.2 Monitored by ESI-MS.
Native-state exchange was carried out by diluting 15 μl of deuterated protein (approximately 700 μM protein concentration) in D2O-buffer, 15-fold into 210 μl of labeling buffer (20 mM sodium phosphate pH 7.2 in H2O) containing 0 M, 0.55 M, or 1.07 M GdnHCl so that the final concentration of GdnHCl was 0, 0.5, or 1.0 M. At different times of labeling, exchange was quenched by decreasing the pH to 2.6, with the addition of an equal volume (225 μl) of quench buffer (100 mM glycine pH 2.3 in H2O). The samples were desalted and analyzed by mass spectrometry (for details, see SI Methods).
Pepsin Digestion.
After HX, quenching and desalting, samples were subject to partial pepsin digestion, by passing them through a column packed with pepsin-agarose beads (from Sigma). The column was equilibrated with water at pH 2.6. For details, see SI Methods.
Data Acquisition by ESI-MS.
A Micromass Q-TOF Ultima mass spectrometer was used to analyze both intact and digested samples. The mass spectrometer was operated in the positive ion mode. The concentration of protein in the samples was typically 11 μM. For details, see SI Methods.
Data Analysis.
MassLynx software was used to determine the widths of mass spectra. Details of data analysis are presented in SI Methods.
Supplementary Material
Acknowledgments.
We thank members of our laboratory for discussions. J.B.U. is the recipient of a J.C. Bose National Fellowship from the Government of India. This work was funded by the Tata Institute of Fundamental Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908617106/DCSupplemental.
References
- 1.Linderström-Lang K, Schellman JA. The Enzymes. 2nd Ed. Vol. 1. New York: Academic; 1959. Protein structure and enzyme activity; pp. 443–510. [Google Scholar]
- 2.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 3.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 5.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 6.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev (Washington, DC) 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 7.Saxena AM, Udgaonkar JB, Krishnamoorthy G. Protein dynamics control proton transfer from bulk solvent to protein interior: A case study with a green fluorescent protein. Protein Sci. 2005;14:1787–1799. doi: 10.1110/ps.051391205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cremades N, Sancho J, Freire E. The native-state ensemble of proteins provides clues for folding, misfolding and function. Trends Biochem Sci. 2006;31:494–496. doi: 10.1016/j.tibs.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Frank H, Franks F. Structural approach to the solvent power of water for hydrocarbons; urea as a structure breaker. J Chem Phys. 1968;48:4746–4757. [Google Scholar]
- 10.Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 11.Robinson DR, Jencks WP. The effect of compounds of the urea-guanidinium class on the activity coefficient of acetyltetraglycine ethyl ester and related compounds. J Am Chem Soc. 1965;87:2462–2470. doi: 10.1021/ja01089a028. [DOI] [PubMed] [Google Scholar]
- 12.Makhatadze GI, Privalov PL. Protein interactions with urea and guanidinium chloride. A calorimetric study. J Mol Biol. 1992;226:491–505. doi: 10.1016/0022-2836(92)90963-k. [DOI] [PubMed] [Google Scholar]
- 13.Lim KW, Rosgen J, Englander SW. Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. Proc Natl Acad Sci USA. 2009;106:2595–2600. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadqi M, Casares S, Abril MA, Lopez-Mayorga O, Conejero-Lara F, Freire E. The native state conformational ensemble of the SH3 domain from alpha-spectrin. Biochemistry. 1999;38:8899–8906. doi: 10.1021/bi990413g. [DOI] [PubMed] [Google Scholar]
- 16.Juneja J, Udgaonkar JB. Characterization of the unfolding of ribonuclease a by a pulsed hydrogen exchange study: Evidence for competing pathways for unfolding. Biochemistry. 2002;41:2641–2654. doi: 10.1021/bi011480p. [DOI] [PubMed] [Google Scholar]
- 17.Bhutani N, Udgaonkar JB. Folding subdomains of thioredoxin characterized by native-state hydrogen exchange. Protein Sci. 2003;12:1719–1731. doi: 10.1110/ps.0239503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrington CB, Teesch LM, Robertson AD. Defining protein ensembles with native-state NH exchange: Kinetics of interconversion and cooperative units from combined NMR and MS analysis. J Mol Biol. 1999;285:1265–1275. doi: 10.1006/jmbi.1998.2338. [DOI] [PubMed] [Google Scholar]
- 19.Arrington CB, Robertson AD. Microsecond to minute dynamics revealed by EX1-type hydrogen exchange at nearly every backbone hydrogen bond in a native protein. J Mol Biol. 2000;296:1307–1317. doi: 10.1006/jmbi.2000.3536. [DOI] [PubMed] [Google Scholar]
- 20.Hoang L, Bedard S, Krishna MM, Lin Y, Englander SW. Cytochrome c folding pathway: Kinetic native-state hydrogen exchange. Proc Natl Acad Sci USA. 2002;99:12173–12178. doi: 10.1073/pnas.152439199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viguera AR, Serrano L, Wilmanns M. Different folding transition states may result in the same native structure. Nat Struct Biol. 1996;3:874–880. doi: 10.1038/nsb1096-874. [DOI] [PubMed] [Google Scholar]
- 22.Northey JG, Di Nardo AA, Davidson AR. Hydrophobic core packing in the SH3 domain folding transition state. Nat Struct Biol. 2002;9:126–130. doi: 10.1038/nsb748. [DOI] [PubMed] [Google Scholar]
- 23.Korzhnev DM, et al. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 24.Borreguero JM, Ding F, Buldyrev SV, Stanley HE, Dokholyan NV. Multiple folding pathways of the SH3 domain. Biophys J. 2004;87:512–533. doi: 10.1529/biophysj.104.039529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wani AH, Udgaonkar JB. Revealing a concealed intermediate that forms after the rate-limiting step of refolding of the SH3 domain of PI3 kinase. J Mol Biol. 2009;378:348–362. doi: 10.1016/j.jmb.2009.01.060. [DOI] [PubMed] [Google Scholar]
- 26.Engen JR, Smithgall TE, Gmeiner WH, Smith DL. Identification and localization of slow, natural, cooperative unfolding in the hematopoietic cell kinase SH3 domain by amide hydrogen exchange and mass spectrometry. Biochemistry. 1997;36:14384–14391. doi: 10.1021/bi971635m. [DOI] [PubMed] [Google Scholar]
- 27.Wales TE, Engen JR. Partial unfolding of diverse SH3 domains on a wide timescale. J Mol Biol. 2006;357:1592–1604. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- 28.Jackson SE. How do small single-domain proteins fold? Fold Des. 1998;3:R81–91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 29.Jaswal SS, Miranker AD. Scope and utility of hydrogen exchange as a tool for mapping landscapes. Protein Sci. 2007;16:2378–2390. doi: 10.1110/ps.072994207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferraro DM, Lazo ND, Robertson AD. EX1 hydrogen exchange and protein folding. Biochemistry. 2004;43:587–594. doi: 10.1021/bi035943y. [DOI] [PubMed] [Google Scholar]
- 31.Udgaonkar JB. Multiple routes and structural heterogeneity in protein folding. Annu Rev Biophys. 2008;37:489–510. doi: 10.1146/annurev.biophys.37.032807.125920. [DOI] [PubMed] [Google Scholar]
- 32.Arrington CB, Robertson AD. Correlated motions in native proteins from MS analysis of NH exchange: Evidence for a manifold of unfolding reactions in ovomucoid third domain. J Mol Biol. 2000;300:221–232. doi: 10.1006/jmbi.2000.3859. [DOI] [PubMed] [Google Scholar]
- 33.Sagle LB, Zimmermann J, Dawson PE, Romesberg FE. Direct and high resolution characterization of cytochrome c equilibrium unfolding. J Am Chem Soc. 2006;128:14232–14233. doi: 10.1021/ja065179d. [DOI] [PubMed] [Google Scholar]
- 34.Bollen YJM, Kamphuis MB, van Mierlo CPM. The folding energy landscape of apoflavodoxin is rugged: Hydrogen exchange reveals non-productive misfolded intermediates. Proc Natl Acad Sci USA. 2006;103:4095–4100. doi: 10.1073/pnas.0509133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason PE, Brady JW, Neilson GW, Dempsey CE. The interaction of guanidinium ions with a model peptide. Biophys J. 2007;93:L04–L06. doi: 10.1529/biophysj.107.108290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennion BJ, Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc Natl Acad Sci USA. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hua L, Zhou R, Thirumalai D, Berne BJ. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc Natl Acad Sci USA. 2008;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stumpe MC, Grubmuller H. Urea impedes the hydrophobic collapse of partially unfolded proteins. Biophys J. 2009;96:3744–3752. doi: 10.1016/j.bpj.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright CF, Lindroff-Larsen K, Randles LG, Clarke J. Parallel protein-unfolding pathways revealed and mapped. Nat Struct Biol. 2003;10:658–662. doi: 10.1038/nsb947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.