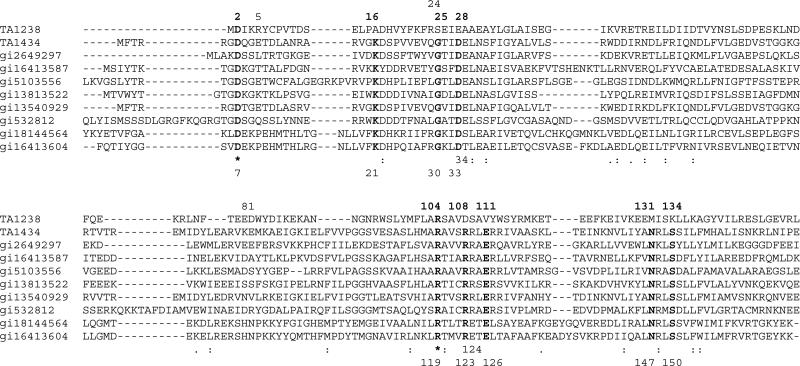
Figure 4.
Sequence alignment of Pfam01923 proteins and TA1238. The proteins are from the following organisms: TA1238 from Thermoplasma acidophilum, TA1434 from Thermoplasma acidophilum, gi2649297 from Archaeoglobus fulgidus, gi16413587 from Listeria innocua, gi5103556 from Aeropyrum pernix, gi13813522 from Sulfolobus solfataricus, gi13540929 from Thermoplasma volcanium, gi532812 from Caenorhabditis elegans, gi18144564 from Clostridium perfringens, and gi16413604 from Listeria innocua. First, Pfam proteins were aligned and then TA1238 sequence was added based on structural alignment of TA1238 and TA1434. Bold letters indicate the residues that are invariant throughout the family, excluding TA1238. Stars show the only two residues that are also invariant in TA1238. Numbers at the top of the alignment are those from the TA1238 sequence and at the bottom – from TA1434. Note residues 34 and 124 in Pfam and 5, 24, and 81 in TA1238, whose possible roles are also discussed in the text.