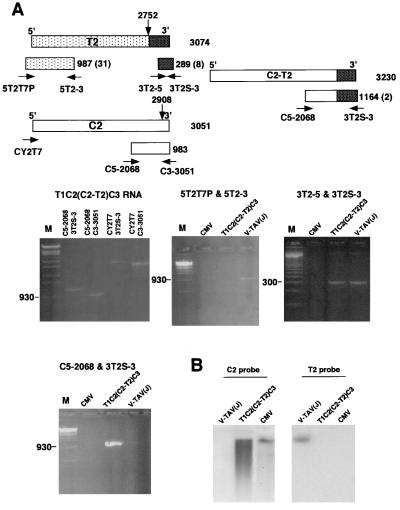
Figure 5.
Detection of C2-T2 and T2 by RT-PCR from the progeny RNAs after T1T2C2C3 inoculation. (A) RT-PCR products using primer pairs shown at top. Primers are: CY2T7 (5′-CCGGATCCATTAATACGACTCACTATAGTTTATTTACAAGAGCG-3′, corresponding to T7 promoter + nt 1–17 of C2), C3–3051 (5′-TGGTCTCCTTTTGGAGGCC-3′, hybridizing to nt 3051–3033 of C2), 5T2–3 (5′-GCTTGTAGTAGATCTACAGTC-3′, hybridizing to nt 956–936 of T2), C5–2068 (5′-GTTCCAGATCCATTGCG-3′, corresponding to nt 2068–2084 of C2), and 3T2–5 (5′-GTCTGAGTTGGTAGTATTGC-3′, corresponding to nt 2793–2812 of T2). The primers of 5T2T7P and 3T2S-3 have been described. The estimated sizes of the PCR products are indicated at the right of each box representing the PCR products. The number of nonviral bases derived from the primers is indicated in parentheses. The cross-hatched box represents the 3′-terminal region that was fused to C2. Marker lanes (M) contain a DNA ladder of StyI-digested λ DNA or a 100-bp DNA ladder (Right, middle row). (Left, middle row) RT-PCR products that were amplified from T1C2(C2-T2)C3 RNAs prepared from the purified virus with the primer pairs indicated for each lane. The three remaining figures show RT-PCR products that were amplified from CMV, T1T2(C2-T2)C3, and V-TAV(J) RNAs by using the primer pairs as indicated. (B) Northern blot analysis of C2 and T2. Extracted RNAs from the purified viruses were subjected to electrophoresis in a 1.5% denaturing agarose gel, blotted onto a nitrocellulose membrane, and hybridized either with C2 probe or T2 probe.