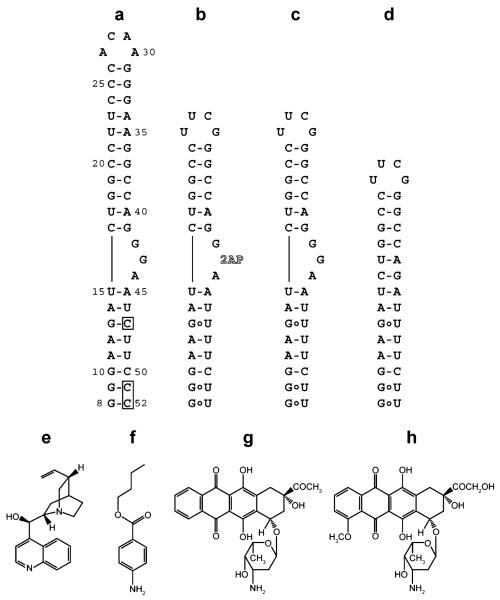
Figure 1.
RNA constructs and selected compounds used in this study. a) HIV-1 frameshift site RNA stem-loop. Boxes indicate where the native uridines at positions 47, 51, and 52 have been changed to cytidines for NMR studies (HIV-18-52). The construct is numbered starting from the first residue of the UUUUUUA slippery sequence (not shown), which is immediately adjacent to the 5′ end of the stem-loop. b) Frameshift site RNA with a truncated upper stem, a native lower helix, and a 2-aminopurine substituted for the central guanosine in the three-purine bulge (HIV-1screen). c) Sequence as in panel b, except the central guanosine remains unaltered (HIV-1wtbulge). d) Sequence as in panel c, except the three-purine bulge has been deleted (HIV-1Δbulge). e) Cinchonidine. f) Butamben. g) Idarubicin. h) Doxorubicin.