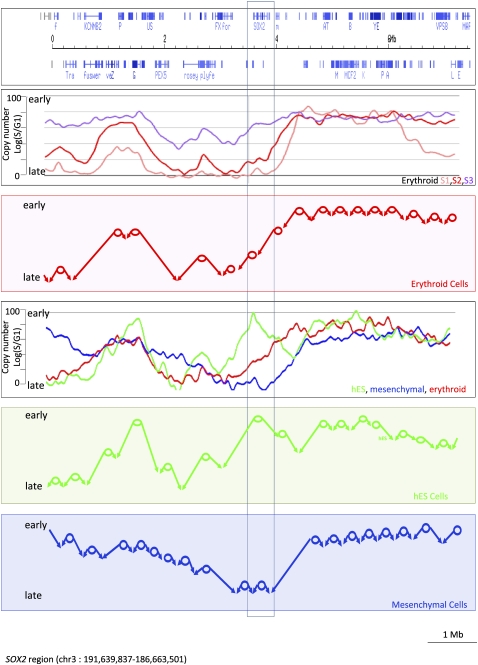
Figure 6.
Model of DNA replication in mammalian cells. (Top panel) An 8-Mb region centered on the SOX2 gene region; the second panel is the result of a TimEX analysis of sorted early, middle, and late S fractions from basophilic erythroblasts. The red circles in the third panel represent the major zones containing origins of replication that can be deduced from an analysis of the second panel; the arrows show the predicted fork direction in erythroid cells starting from these origins. The main features of the model are that (1) active origins of replication are unevenly distributed in the genome and relatively rare in parts of the genome, creating large initiation free regions, (2) origins are programmed to fire within narrow time windows during S phase, (3) lack of pause sites creates long transition regions in which the forks progress unidirectionally. The fourth panel represents the TimEX profiles of whole S fractions from basophilic erythroblasts (red), mesenchymal cells (blue), and hESC (green) in the same region. The two bottom panels illustrate the predicted organization of the replication of this genomic segment in hESC and mesenchymal cells. One segment with major differences in timing between the three cell lines is boxed in the immediate neighborhood of the SOX2 gene, which is expressed at a high level in hESC and silent in erythroid and mesenchymal cells (data not shown). Because of the paucity of active origins in the region, the tissue-specific activation in hESC of an early origin or zone of initiation, which appears to coincide with the SOX2 gene, has dramatic repercussions in both the timing of replication and the fork directions in a megabase-wide region.