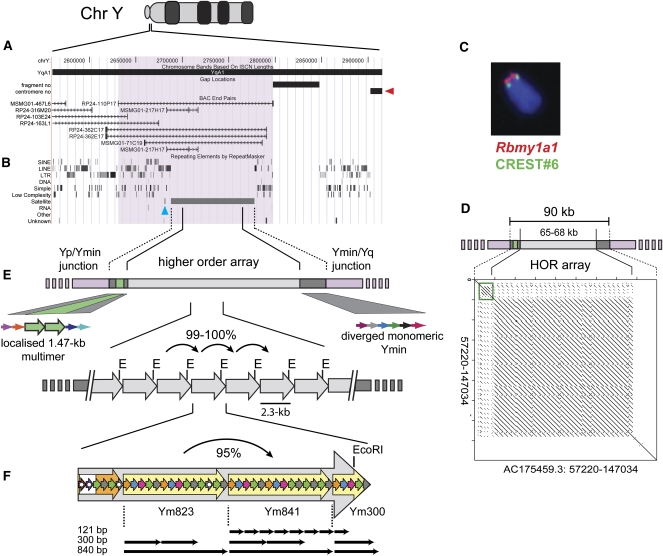
Figure 1.
Structural and sequence map spanning the C57BL/6J mouse Y centromere. (A) UCSC mouse Genome Browser view (July 2007, Build 37 assembly) at the unmapped Y centromere. BAC 110P17 (coverage highlighted mauve) is the most proximally annotated BAC to the unanchored Y centromere (arbitrarily indicated at right, red arrowhead). (B) Repeat elements identified by RepeatMasker show Ymin as a 90-kb tract of uninterrupted satellite DNA except for a single, mouse-specific LINE1 element (L1Md_F) that isolates ∼620 bp of Ymin (blue arrowhead) at the 5′ edge of the array. The most closely annotated gene, a multicopy RNA-binding motif protein (Rbmy1a1), lies ∼290 kb on the short arm-side of the Y centromere, as shown in (C) by immuno-FISH using Rbmy1a1 DNA FISH probe (red) and CREST#6 anti-centromere antibody (green). (D) Self-alignment dot-plot of Ymin reveals a 65–68-kb tandem array of 2.3-kb HOR units flanked by 10–12 kb of diverged, localized multimeric (green box) and monomeric Ymin. (E) HOR periodicity of 2.3 kb is also indicated by regularly-spaced restriction sites (e.g., E, EcoRI; see Supplemental Fig. S1) within the homogeneous Ymin array, with individual units sharing 99%–100% sequence identity. (F) Each canonical HOR unit comprises 2.4 copies of an ∼840-bp periodicity subunit repeat (large yellow arrows designated Ym823, Ym841, and Ym300; see Supplemental Fig. S2) and a less uniform region lacking repeat periodicity or with 60-bp periodicity (large composite white/orange arrow, respectively). Highly diverged ∼60–61-bp monomers (small colored arrows representing monomers with similar homology) or 121-bp monomers are arranged in HOR subunits with periodicities of ∼300–840 bp (examples shown by black arrows). No subunit repeat extends the full length of the HOR unit domain (Supplemental Table S3A). Monomers harboring deletions or other rearrangement are highlighted by solid white circles.