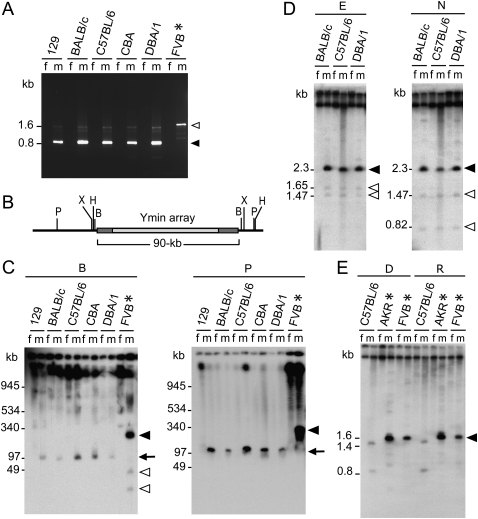
Figure 2.
Molecular characterization of Ymin in inbred mice. (A) Ymin PCR assay amplifies male (m) but not female (f) paired genomic DNA samples and differentiates the Y centromeres of M.m. musculus (molossinus) (0.82 kb) and M.m. domesticus (1.6 kb) (also see Supplemental Fig. S4). (B) Restriction map of Ymin domain showing enzymes BamHI (B), HindIII (H), PacI (P), and XbaI (X) predicted to release the 90-kb Ymin array. (C) PFGE Southern hybridization of genomic DNAs probed with Ymin exhibits male-specific fragment lengths of predicted size (arrow) in five strains carrying a M.m. molossinus Y chromosome after digestion with enzymes BamHI (90.7 kb expected) and PacI (124 kb expected) (HindIII and XbaI data not shown; also see Supplemental Fig. S6). Strain FVB (asterisk), carries a M.m. domesticus Y chromosome with an array length of 325–350 kb (sum total of fragments). Additional, smaller fragments (open arrowheads) produced by enzymes BamHI (and HindIII data not shown) suggests increased sequence complexity at the flanking edges of the larger M.m. domesticus Ymin array (solid arrowhead). (D) Short-range Southern analysis of Ymin in the same five M.m. molossinus strains (129 and CBA data not shown) following EcoRI (E) and NcoI (N) digestion confirms a HOR unit length of 2.3 kb (solid arrowhead). Predicted rare variant fragment lengths of 1.47 and 1.65 kb (EcoRI) and 0.82 and 1.47 kb (NcoI) are seen as faint hybridization bands (open arrowheads). (E) Mice carrying a M.m. domesticus Y chromosome (AKR and FVB, *) reveal a Ymin HOR unit length of 1.6 kb following digestion of genomic DNA with DraI (D), RsaI (R), and TaqI (data not shown). The C57BL/6 (molossinus) Ymin HOR cleaves twice, as predicted by BAC 110P17 sequence data.