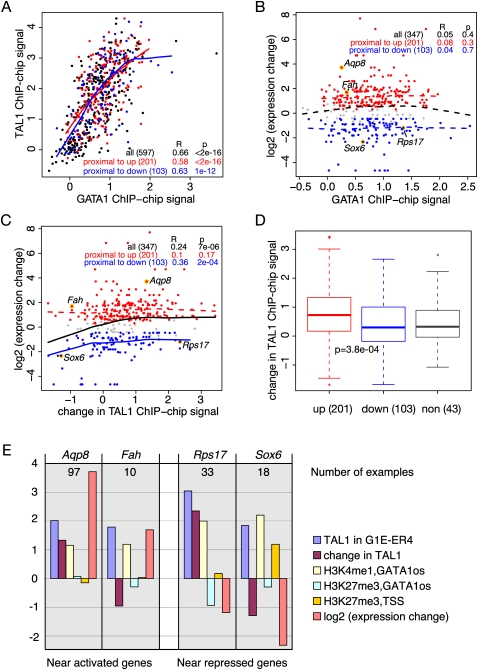
Figure 4.
Correlation among GATA1, TAL1, and changes in gene expression. (A) Scatterplot for the level of occupancy by GATA1 (x-axis) and TAL1 (y-axis) for all GATA1 OSs in the 66-Mb region of mouse chromosome 7, with the GATA1 OSs in the proximal neighborhood of up-regulated genes shown as red dots (increase in expression above the FDR threshold of 0.001), those in the proximal neighborhood of down-regulated genes shown as blue dots (decrease in expression exceeding the FDR threshold of 0.001), and all others as black dots. The ChIP-chip hybridization levels for the several probes in each interval covering a GATA1 OS were averaged and used as a proxy for occupancy for GATA1 and TAL1. The Pearson's correlation coefficient R and P-value are listed for three categories of GATA1 OS (all, and those in the proximal neighborhoods of up- or down-regulated genes), and the lowess line is drawn separately for each category of GATA1 OSs. The lowess lines for nonsignificant associations are broken whereas those for significant associations are solid. (B,C) Scatterplots for the relationship between the change in expression level for the genes in whose proximal neighborhood a GATA1 OS is found (y-axis) and GATA1 occupancy (x-axis in B) or the change in TAL1 occupancy between G1E-ER4 cells (GATA1 restored and activated) and G1E Gata1 knockout cells (x-axis in C). The largest difference in expression level (compared with that at time 0) at any point in the time course after activation is the expression change. (D) Boxplots of the distributions of values for the change in TAL1 occupancy in GATA1 OSs associated with genes in three expression categories (non, nonresponsive; up, induced; down, repressed). The differences between the up- and down-regulated categories are significant by a Student's t-test (P = 3.3 × 10−5). (E) Examples illustrating the range of features observed at GATA1 OSs in the neighborhoods of two induced genes and two repressed genes (right). The bars are the mean ChIP-chip signals for the probes in the interval for each GATA1 OS (see key) or the log (base 2) of the expression change. The number of GATA1 OSs that fit the pattern shown is given in each graph (see Table 1).