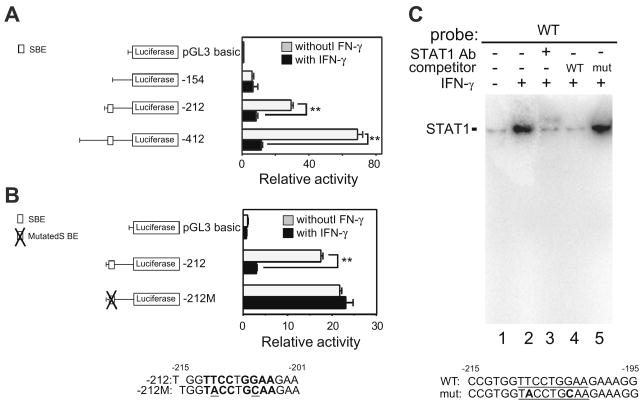
Fig. 3.
Identification of an IFN-γ-responsive region in the SCGB3A1 gene promoter. (A) Transfection analysis of a series of luciferase reporter plasmids containing various lengths (–412, –212 and –154/+52 bp) of the mouse SCGB3A1 gene promoter with or without IFN-γ. The proximal SBE (5′-TTCCTGGAA-3; –212 to –154 bp) previously shown to be involved in the IL-4/IL-13-induced increase of SCGB3A1 expression[15], is indicated in the illustration on the left. The SBE is characterized by a palindromic sequence shown in boldface. Luciferase assays were performed in mtCC cells with (black bar) or without (gray bar) stimulation with 100 ng/ml of IFN-γ. Results shown are the mean ± SE from three independent experiments. A significant difference was obtained with versus without IFN-γ treatment: **P < 0.01. (B) Mutation analysis. The SBE site and the mutated sequences (underlined) are shown in boldface. The mutation of the proximal SBE abrogates the IFN-γ-induced promoter activity of the –212 construct. (C) Electrophoretic mobility shift assays (EMSA) for the STAT1-binding site in the SCGB3A1 gene promoter. A SBE is underlined. In the mutated SBE oligonucleotide (mut), mutations were introduced by changing 2 nucleotides (boldfaced). Nuclear extract proteins (5 μg) from RAW264.7 cells stimulated with 100 ng/ml of IFN-γ for 30 min at 37 °C were incubated with labeled wild-type (WT) SBE oligonucleotide. For supershift assays, 2 μg of STAT1 antibody was added to probe-extract complexes. After incubations, DNA/protein complexes were resolved by 6% polyacrylamide gel electrophoresis.