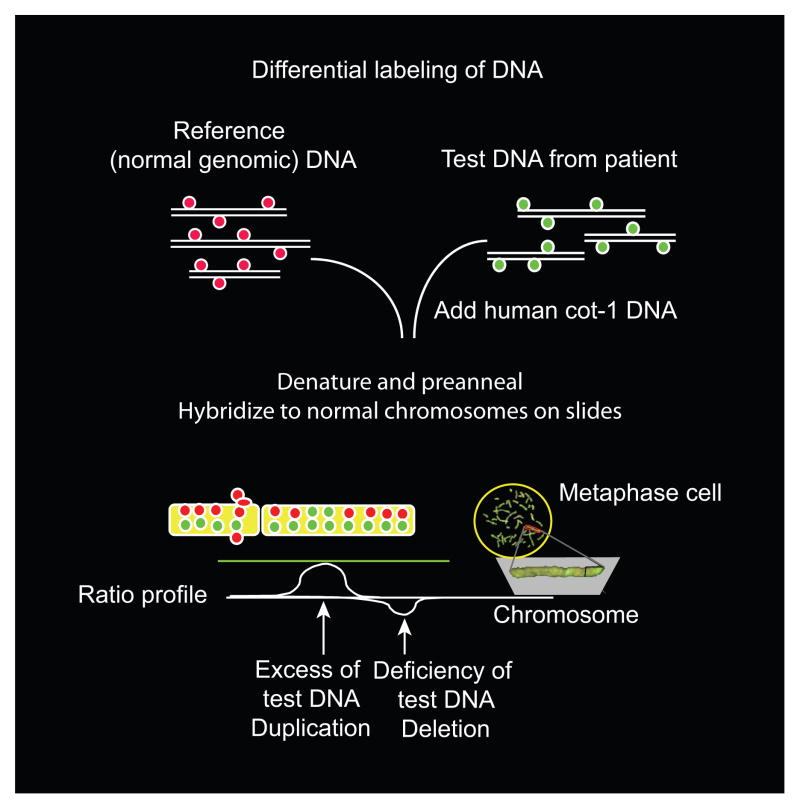
Fig. 2.
The potential use of comparative genomic hybridization to diagnose structural genomic changes in stillbirth. This figure illustrates the general principle underlying comparative genomic hybridization, which is used to identify DNA deletions or duplications. In this technique, fluorescently labeled DNA from a normal control sample is mixed with a test sample that is labeled with a different-colored dye. The mixture then is hybridized to normal metaphase chromosomes. Regional differences in the fluorescence ratio represent gains and losses in the sample DNA compared with the control DNA. Arrays containing hundreds of thousands of small, well-defined DNA probes have replaced normal metaphase chromosomes, allowing more precise and detailed analysis of small, underrepresented or overrepresented areas. Figure courtesy of Dr. Ronald Wapner.
Reddy. Stillbirth Classification of Cause of Death. Obstet Gynecol 2009.