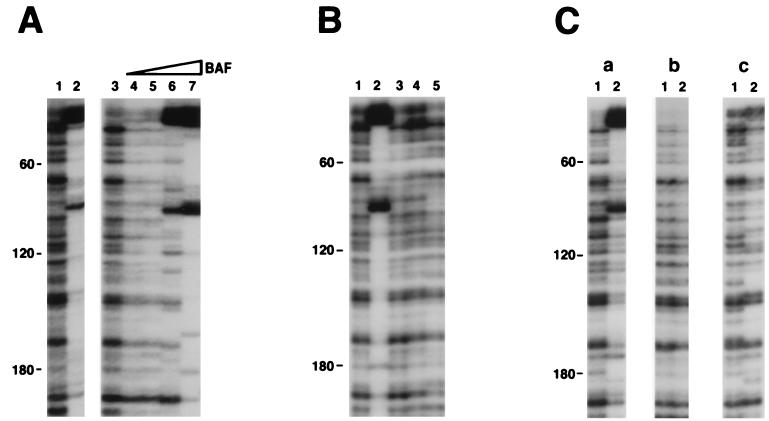
Figure 1.
Reconstitution of the footprints on the viral DNA U3 end. All preintegration complexes were treated with 750 mM KCl before separation from free proteins on a 10–50% Nycodenz gradient. Reconstitution reactions then were carried out on the gradient purified complexes, followed by MM-PCR footprinting (see Materials and Methods for details). End-labeled primer SW2 (13) was used for PCR. Numbers on the left side of each panel indicate the distance from the very end of the viral DNA. Distinctive enhancements are located at ≈20 bp and ≈100 bp from the U3 end. (A) Reconstitution with BAF protein. Samples are naked viral DNA (lane 1) and MLV preintegration complexes (lane 2). Reconstitution reactions included: lane 3, buffer only control, and lanes 4–7, BAF (0.2, 0.6, 2, and 6 ng, respectively). (B) Reconstitution with various protein factors. Reconstitution reactions included: lane 1, buffer only control; lane 2, BAF (2 ng); lane 3, murine HMG Y; lane 4, human HMG I(Y); and lane 5, human HMG1. Purified protein factors shown in lanes 3–5 have been tested in a range of 0.5 to 5 μg, and no differences were observed in this range (data not shown); therefore only reactions with 1 μg of protein are shown. (C) Enhancements are observed only with complexes containing integrase. Reconstitution reactions with BAF were carried out with (a) salt-stripped MLV preintegration complexes, (b) naked viral DNA, and (c) salt-stripped integrase-minus MLV preintegration complexes. Lanes 1 and 2 included 0 and 2 ng BAF, respectively.