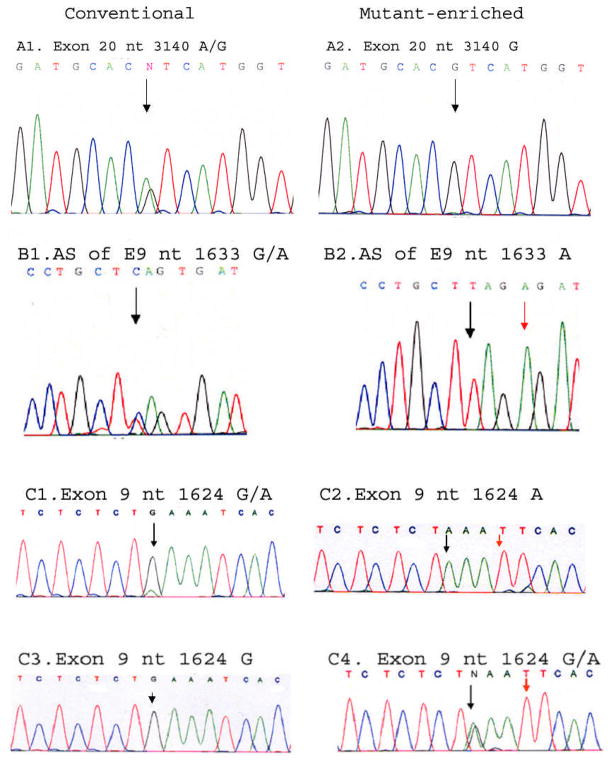
Fig. 3. The detection of PIK3CA hot-spot mutations by mutant-enriched sequencing.
A1–2. The sensitivity of the mutant-enriched sequencing protocol for the exon 20 H1047R mutation was investigated in head and neck cell line Detroit 562, whose genome had been reported to harbor a H1047R mutation 11. A1. Both wild-type A and mutant G peaks were detected at 1:1 ratio as expected by conventional genomic sequencing of the cell line Detroit 562 DNA (non-diluted). A2. When the ratio of mutant and wild-type DNA reached 1: 360, the mutant G peak was still the only peak detected in the cell line Detroit 562 DNA by mutant-enriched sequencing. B1–2. A patient sample with a known E545K mutation was used to test the mutant-enriched sequencing protocol for the exon 9 E545K mutation. B1. Antisense sequencing of the mutated site by the conventional genomic sequencing (marked by the black arrow) detected both the wild-type and the mutant alleles. B2. The peak representing the wild-type allele in the same sample disappeared when the mutant-enriched sequencing method was applied (indicated by the black arrow). The red arrow marks the nucleotide A1630T change that was introduced by mismatch primer (PIK-E9MF) in order to generate the restriction enzyme Hpy188I (TCNGA) site. C1–4. The mutant-enriched sequencing method identified an undetected PIK3CA hotspot mutation E542K (G1624A) by the conventional sequencing. C1. Forward sequencing of a clinical sample with a known E542K mutation by the conventional genomic sequencing method (marked by the black arrow) displayed a dominant presence of the wild-type allele over the mutant allele. C2. The wild-type allele in the same sample disappeared when the mutant-enriched sequencing method was applied (indicated by the black arrow). The red arrow marks the nucleotide A1627T change that was introduced by mismatch primer (PIK-2E9MF) to generate a unique restriction enzyme EcoRI (GAATTC) site. The case of pharyngeal cancer that had been tested negative for a mutation by the conventional genomic sequencing method (C3), but was subsequently identified with a PIK3CA E542K mutation using the mutant-enriched sequencing method (C4).