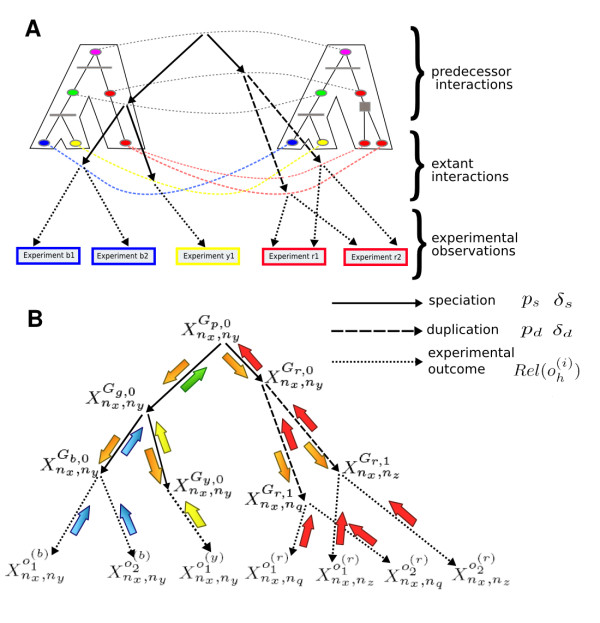
Figure 8.
Inferring protein interactions via message passing. A toy example of the Bayesian tree model of evolution of interactions between members of two protein families for three species: blue, yellow and red. For each species a certain number of experimental datasets is given: two for blue and red and one for yellow. Part (A) shows two reconciled trees for the considered families together with putative protein interactions at each level of evolution. The proteins in the trees are represented by ellipses (with color corresponding to their species). The speciation events are marked by horizontal lines and the duplication events are marked by filled squares. The evolution of the ancestral interaction between the root proteins (purple) can be traced down the trees to the extant interactions. Evidence for the extant interactions can be found in the experimental datasets. In (B) a random variable is associated with each putative interaction. A solid arrow indicates a dependence between two random variables which comes from the speciation event. Similarly, a dashed arrow indicates a dependence for the duplication event. Finally, dotted arrows represent an interface between the true interactions in extant species and the observed experimental evidence. The parameters ps, δs, pd and δd determine the probability of retaining or gaining an interaction during evolution, while the reliability of each dataset (Rel(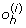 )) determines the probability of identifying a true interaction or a false positive one. Arrows colored blue, yellow, red and green represent messages corresponding to interaction evidence coming from each of the species. These messages are passed up the tree in the first phase of the MP algorithm. In the second phase, messages containing aggregated evidence from one side of the tree are passed down to the other side (orange arrows).
)) determines the probability of identifying a true interaction or a false positive one. Arrows colored blue, yellow, red and green represent messages corresponding to interaction evidence coming from each of the species. These messages are passed up the tree in the first phase of the MP algorithm. In the second phase, messages containing aggregated evidence from one side of the tree are passed down to the other side (orange arrows).