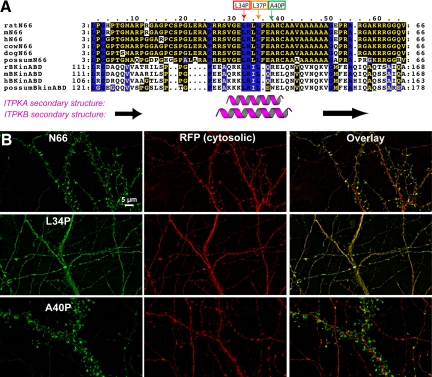
Figure 2.
The L34P point mutation within the ITPKA F-actin–binding domain destroys its localization to dendritic spines. (A) Alignment of F-actin–binding domains of ITPKA (top six sequences) and ITPKB (bottom four sequences) from various mammals. Residues shaded in yellow with a black background are identical; black residues with a blue background are conserved among all sequences shown. The cartoons under the alignment depict the consensus of a protein secondary structure prediction program (JPred3). The purple helices and black arrows demarcate predicted α-helices and β-strands, respectively. The small boxes and arrows above the alignment indicate three residues within the predicted, conserved helix. These residues were each singly mutated to proline to test for their effects on F-actin localization in cells. (B) Effects of point mutations on localization of N66 in hippocampal neurons. Plasmids coding for N66, N66(L34P), or N66 (A40P) fused at their C-terminus to mEGFP were each were cotransfected with the soluble marker tdTomato into primary hippocampal neurons at 8 DIV and fixed at 12 DIV. Nonmutated N66 is highly concentrated in F-actin–rich dendritic spines (top). The L34P mutation rendered the GFP fluorescence coincident with the soluble marker (middle). By contrast, the A40P mutation (bottom) had no obvious effect on the targeting, as GFP fluorescence remained highly associated with dendritic spines, similar to the unmutated N66 region. The L37P mutation produced a phenotype intermediate between the two other mutants (not shown). Scale bar, 5 μm.