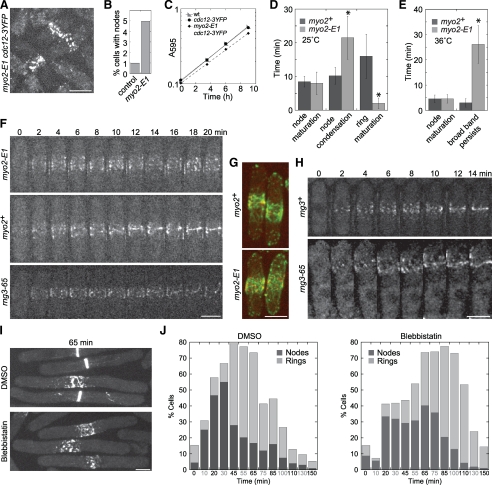
Figure 6.
The myosin-II motor activity is essential for condensation of the broad band of nodes into a contractile ring. (A and B) Localization of Cdc12p-3YFP to the nodes in control myo2+ and myo2-E1 mutant at the permissive temperature of 25°C. Cells (n > 1000 cells each strain) before or at any stage of node condensation were counted as long as nodes could be distinguished. (C) Growth rate of myo2-E1 and myo2+ strains at 25°C. The Y-axis is a logarithmic scale. (A–C) Strains JW81, JW1404, and JW1423 were used. (D–I) The myosin-II motor activity is required for condensation of nodes into a contractile ring. (D and E) Cells expressing both Rlc1p-tdTomato and GFP-CHD in myo2+ (JW1349) and myo2-E1 (JW1701) were imaged at 25 or 36°C. Cytokinesis events were defined as the following: Node maturation, the time between the appearance of Rlc1p nodes and that of actin filaments in the broad band; Node condensation, the time from actin appearance to formation of a compact ring with no lagging nodes; Ring maturation, the time from a compact ring to the beginning of ring constriction; Broad band persists, the time that the broad band of nodes remains uncondensed after the appearance of actin filaments among nodes until the end of 40-min movies. Number of cells analyzed (left to right): (D) 11, 28, 10, 26, 10, and 45; (E) 9, 12, 8, and 22. *p < 0.005. (F) Representative cells from the data in E as well as rng3-65 (JW1720) imaged at 36°C. Only the Rlc1p-tdTomato signal is shown for clarity. The signal is weaker at 36°C than at 25°C, so nodes are somewhat harder to visualize. (G) GFP-CHD–labeled actin filaments associate with Rlc1p-labeled nodes. Maximum intensity projection (seven sections spaced at 0.8 μm) of cells expressing both Rlc1p-tdTomato (red) and GFP-CHD (green) in myo2+ (top, JW1349) or myo2-E1 mutant (bottom, JW1701). (H) The contractile ring assembles from nodes after the recovery of Myo2p motor activity. Cells of rng3+ (JW1666) or rng3-65 (JW1720) were shifted to 36°C for 4 h and then back to 24°C for 4 h before imaging at 24°C. (I and J) cdc25-22 rlc1-tdTomato cells (JW1351) were synchronized at 36°C for 4 h and then treated with 1 mM blebbistatin or an equal volume of DMSO at 36°C for 30 min before releasing to 25°C for imaging at time zero. (I) Representative DMSO and blebbistatin-treated cells at 65 min after release. Maximum intensity projection of eight slices spaced 0.6 μm apart. (J) Quantification of cells in each culture that have Rlc1p nodes (dark) and rings (light) at each time point (n > 45 cells for each column). Bars, 5 μm.