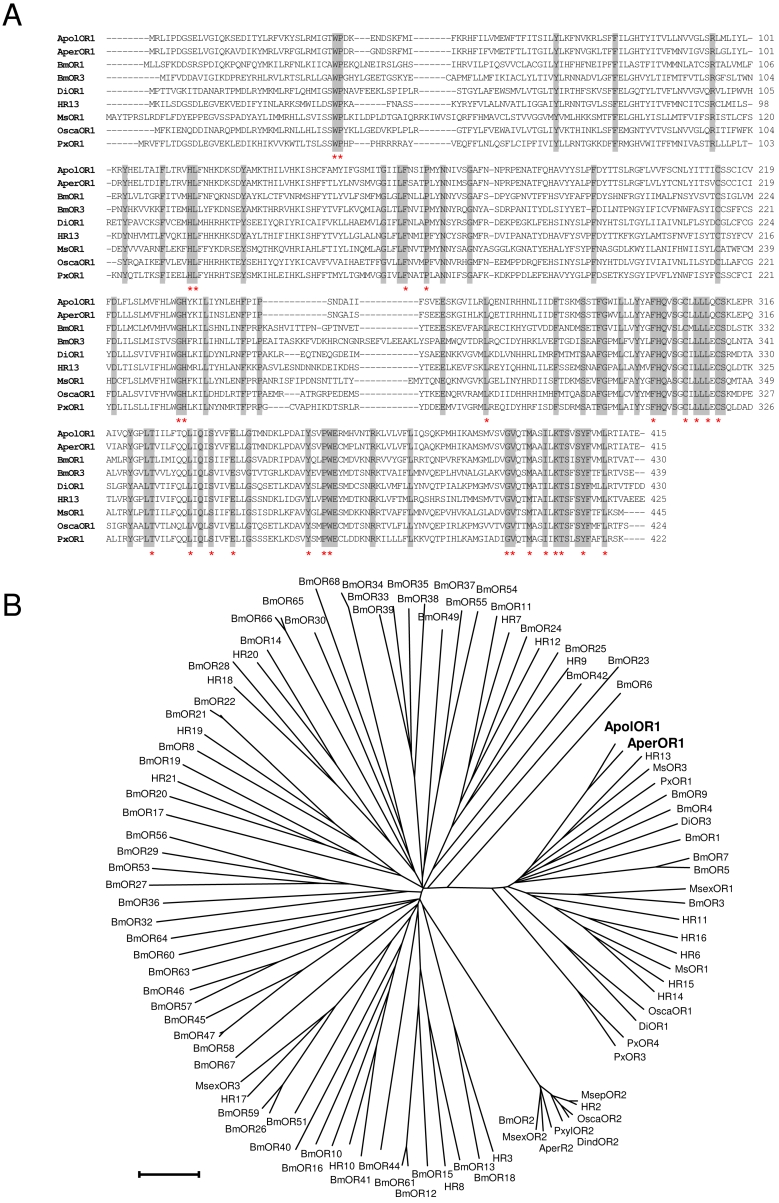
Fig 1.
Sequence comparisons. (A) Alignment of amino acid sequences of ApolOR1, AperOR1 and of selected verified pheromone receptors. Identical amino acids are shaded grey. Amino acids conserved throughout all putative and verified pheromone receptor sequences forming a subfamily with the Antheraea receptors in (B) are indicated by asterisks. (B) Sequence relatedness of moth olfactory receptors. Neighbor joining tree based on the identity between olfactory receptors of different moth species. The distance tree was calculated using the MEGA program and is based on a ClustAL alignment of the sequences indicated. Branch lengths are proportional to percentage sequence difference. Scale bar: 10% difference.