Figure 2.
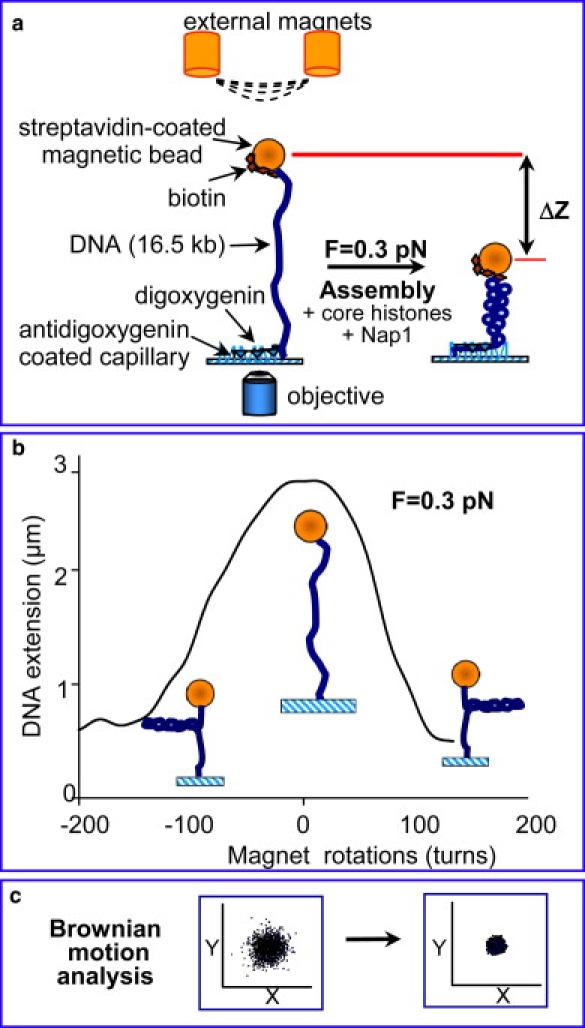
(a) Schematic of the magnetic tweezers setup for studying nucleosome array assembly. A single double-stranded DNA molecule is suspended between a magnetic bead and the surface of the cuvette in a torsionally constrained manner. A mixture of core histones and histone chaperone Nap1 was injected into the cuvette to initiate assembly. The shortening in DNA extension due to formation of nucleosomes was measured by monitoring the movement of the bead in (z), as well as by Brownian motion analysis. (b) Rotation-extension curve on the torsionally constrained DNA at F = 0.3 pN. At low force, rotation of the two external magnets induced (+) or (−) supercoiling in the DNA molecule, which led to plectoneme formation, and thus shortening of the DNA tether. (c) Brownian motion scatter in (x,y) for the naked, relaxed DNA was larger than for the supercoiled DNA or reconstituted nucleosomal array.
