Figure 6. CD4 silencing negatively modulates CCR5 surface expression in THP-1 and primary T cells.
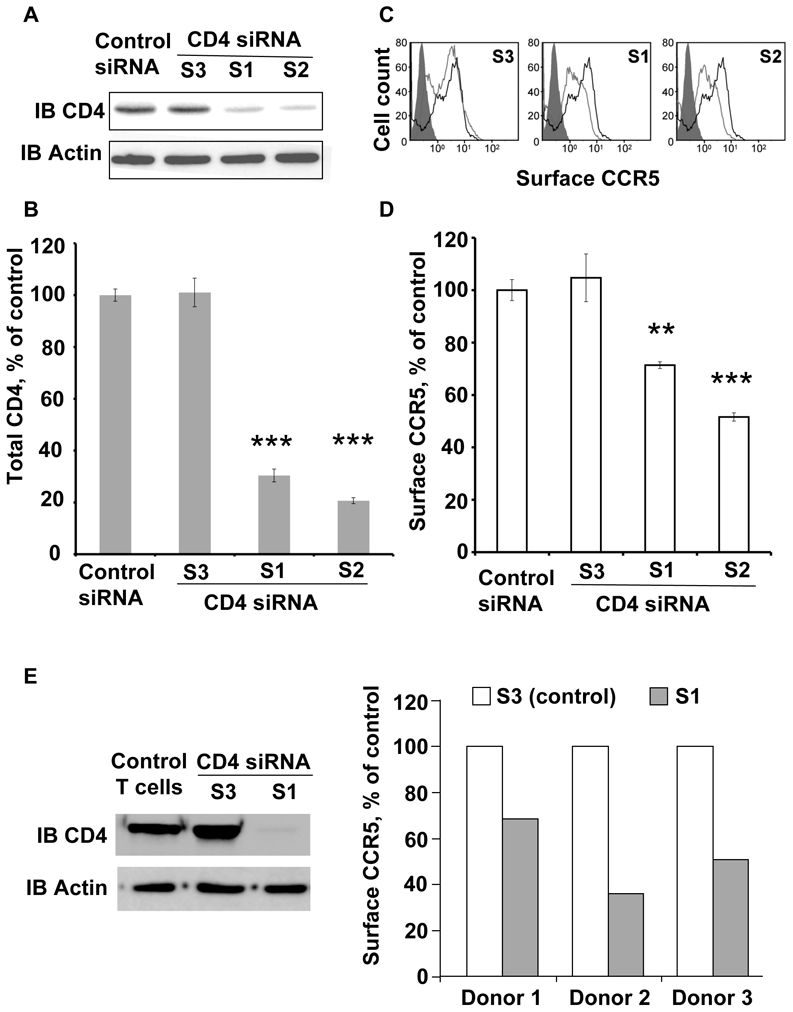
(A) Western blot analysis of CD4 from THP-1 cells nucleofected with siRNAs directed against CD4 (S1, S2 and S3) or scrambled control. CD4 was detected using the 1F6 anti-human CD4 antibody. (B) Quantification of CD4 after treatment with indicated siRNAs (n=3, in triplicates). Asterisks indicate statistical significance (p<0.001) with cells treated with control siRNA. (C) FACS analysis of cell surface CCR5 using the 1/85a Alexa-fluor 647-conjugated anti-human CCR5. Grey open histograms of S1, S2 and S3-treated cells are compared to the control (scramble siRNA) histogram in black. Grey filled histogram corresponds to isotypic control. (D) The effect of CD4 inhibition on surface CCR5 was measured as follows: the proportion of surface CCR5 was calculated by FACS (by comparing the signal in unpermeabilzed and permeabilized cells) in 3 independent experiments, in triplicates (**p<0.01; ***p<0.001); the values were compared to the proportion of surface CCR5 in THP-1 nucleofected with control scramble siRNA. (E) Left panel: Representative western blot analysis of CD4 from donor-purified, activated (see methods) primary T cells, nucleofected with 2 siRNAs targeting CD4 sequences (S1 and S3); note that, as for THP-1 cells, the S3 siRNA did not reduce CD4 concentration in T cells. Right panel: effect of CD4 inhibition on surface CCR5 in T cells from 3 donors. The proportion of surface CCR5 was calculated by FACS, by comparing the signal in unpermeabilzed and permeabilized cells, and values obtained in cells nuleofected with the S1 siRNA were compared to the proportion of surface CCR5 in cells nucleofected with the ineffective S3 siRNA.
