Figure 2.
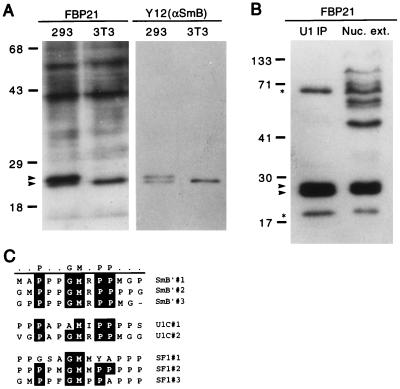
The WW domain of FBP21 binds SmB as determined by blot overlay. (A) Total cell lysate probed with the 32P-GST-FBP21 WW domain. Total cell lysates from 293 cells (a human cell line) and NIH 3T3 cells (a mouse cell line) were prepared in RIPA buffer. Lysates were run on an SDS/PAGE gel, were blotted onto nitrocellulose, and were incubated with radiolabeled GST-FBP21 WW (see Materials and Methods). A duplicate blot was incubated with the Y12 antibody to visualize the spliceosomal proteins, SmB, and its variant SmB′. The blot was developed by using enhanced chemiluminescence (ECL, Amersham). The upper arrow identifies SmB′; the lower arrow identifies SmB. (B) SmB and SmB′ are detected in HeLa cell nuclear extract (Nuc. ext.) and immunoprecipitates of HeLa cell nuclear extract. Immunoprecipitates were performed with an anti-U170 antibody to “pull down” U1-associated proteins (U1 immunoprecipitates), which were resolved side-by-side on a SDS/PAGE gel, were blotted onto nitrocellulose, and were incubated with radiolabeled GST-FBP21 WW. The upper starred band runs in the expected position of SF1, and the lower starred band runs in the position of U1C. Molecular mass markers (in kDa) are indicated. (C) An alignment of SmB′, U1C, and SF1 reveals a motif rich in proline, glycine, and methionine residues. This motif is repeated three times in SmB′ and SF1 (SF1-HL1) and twice in U1C. The alignment was carried out by using the program megalign (DNAstar, Madison, WI).