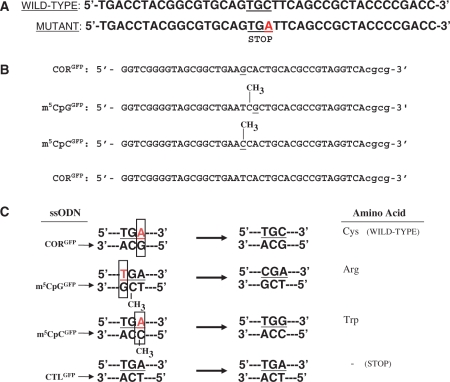
Figure 1.
Wild-type and mutant GFP sequences and ssODN design. (A) The pGFPmut vector contains a single point mutation at base pair +213 of the GFP coding sequence that creates a stop codon in this position. ssODNs were designed to target bases in the stop codon to restore GFP expression. (B) Sequences of targeting (CORGFP, m5CpGGFP, m5CpCGFP) and control (CTLGFP) ssODNs. (C) Annealing of the targeting ssODNs to the genomic sequence creates a mismatch (box), whereas the control ssODN pair perfectly. The CORGFP ssODN is designed to induce an A-to-C conversion that eliminates the stop codon and restores wild-type GFP expression. The m5CpGGFP ssODN creates a G:T mismatch in the context of a m5CpG, thus mimicking the delamination of m5C. This will induce a T-to-C substitution which will convert the stop codon into an arginine. The m5CpCGFP ssODN targets the same base as CORGFP but contains an m5C modification which will lead to an A-to-G conversion and result in a single amino acid substitution (a tryptophan in place of a cysteine). The CTLGFP ssODN perfectly complements the mutant sequence and therefore should not induce any genomic alterations.