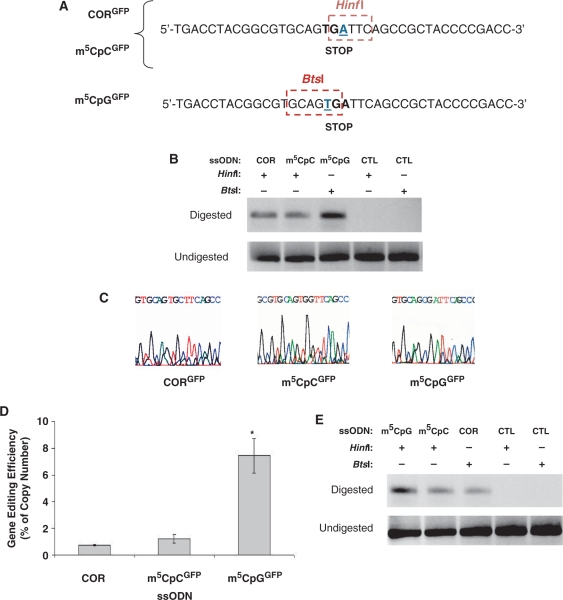
Figure 8.
Evidence of ssODN-mediated single base modification at the molecular level. (A) The single base mutation on the GFP reporter vector (base underlined) generates a new restriction site for HinfI endonuclease. Promotion of an A-to-C transversion mediated by the CORGFP ssODN and an A-to-G transition mediated by the m5CpCGFP ssODN eliminate the restriction site and render the sequence refractory to HinfI digestion and cleavage. The T-to-C conversion mediated by m5CpGGFP eliminates the BtsI restriction site. (B) Genomic DNA was isolated from myoblasts stably expressing the mutant GFP 1 week after ssODN transfection and treated with either HinfI (for CORGFP, m5CpCGFP and CTLGFP) or BtsI (for m5CpGGFP and CTLGFP). Amplification was carried using a forward and a reverse primer encompassing the GFP mutation. No amplification was detected in samples treated with CTLGFP using either HinfI or BtsI. A specific PCR product of identical molecular weight to that obtained after amplification of a wild-type GFP plasmid was detected only in cells treated with targeting ssODNs. (C) Direct sequencing of PCR products obtained as in (B) by amplifying total genomic DNA having undergone HinfI or BtsI digestion demonstrated single base alterations at the genomic level and the specificity of the amplification products. (D) Real-time PCR was used to determine the relative amount of genomic DNA having undergone ssODN-mediated single base alteration. Cultures treated with m5CpGGFP showed 10-fold higher levels of gene editing compared with cultures treated with m5CpCGFP. No significant differences in efficiency were detected between cultures treated with the CORGFP and the m5CpCGFP ssODNs. The total amount of plasmid was determined by amplification of the undigested samples using the same primer combinations and was used as internal standard control to normalize DNA levels. *P = 0.001. (E) Total mRNA was isolated 1 week after ssODN transfection and reverse transcribed using an oligo dT primer. The cDNA was then digested overnight with HinfI or BtsI depending of the ssODN transfected (A) and amplified using the same primers used for the genomic DNA analysis. Modification of the mutant GFP sequence using the m5CpGGFP ssODN was more efficient than that observed in cultures treated with targeting ssODNs lacking the 5-methylcytosine (CORGFP) or the ssODN containing the m5C modification but not in a CpG context (m5CpCGFP).