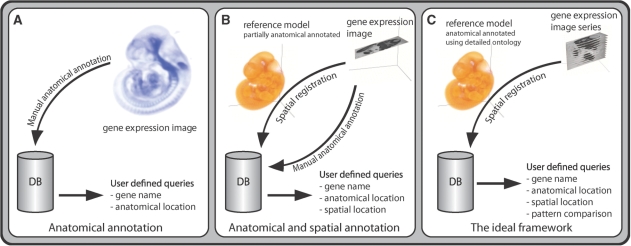
Figure 1.
Frameworks. Panels A and B show the different frameworks used in the discussed atlases. All atlases except for EMAGE use a purely text-based annotation (A). In EMAGE the gene expression images are mapped into reference models (B). The ideal framework is shown in (C). Basically, this framework is used by the Allen Brain Atlas (except for the detailed ontology). The approach involves the mapping of sets of in situ gene expression images to a spatial framework. Enabling automated textual annotation and analysis based on the spatial location of the expression of a gene.