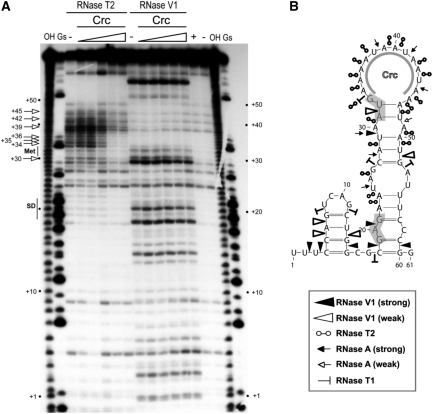
Figure 2.
Secondary structure of the alkS mRNA leader region and effect of Crc binding monitored by enzymatic probing. (A) Fractionation of a 5′-end-labelled alkS mRNA fragment (nts +1 to +78) digested with RNase T2 and RNase V1 on a 12% polyacrylamide–urea gel. The enzymatic digestions were performed in the absence (−) or in the presence of increasing concentrations of Crc (3, 16, 80, 400 nM or 2 µM). Incubation controls (in the absence of RNase) were done in the absence (−) and in the presence (+) of Crc (80 nM). Lanes ‘OH’ and ‘Gs’ correspond, respectively, to an alkaline ladder and an RNase T1 ladder performed under denaturing conditions. Arrows indicated the nucleotides protected by Crc from the attack of RNase T2. Positions relative to PalkS2 transcription start site, as well as the Shine–Dalgarno sequence (SD) and the AUG codon (Met), are indicated on the sides of the gel. (B) Summary of the cleavage patterns generated by RNase V1, RNase T2, RNase A and RNase T1 depicted on the secondary structure model of the alkS mRNA. The symbols used for each RNase are indicated in the insert. The region protected by Crc in the apical loop, as deduced from (A), is indicated. The SD sequence and the AUG start codon are shaded.