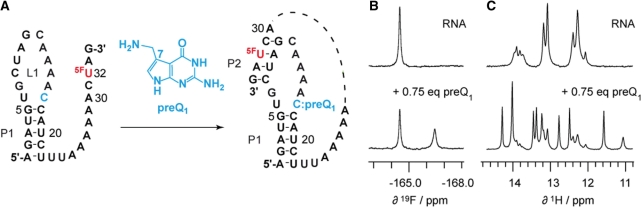
Figure 7.
PreQ1 sensitive riboswitch aptamer. (A) RNA secondary structure model for ligand-induced folding (49). The 5-fluoro uridine label is positioned in a single-stranded region in the free RNA whereas it is located in a double-helical segment in the ligand-bound form. (B) 19F NMR spectra before and after addition of preQ1. (C) Corresponding 1H NMR spectra. Increase of ligand concentration results in higher population of the aptamer/ligand complex (data not shown). Conditions: 0.6 mM oligonucleotide, 25 mM sodium arsenate buffer, H2O/D2O = 9/1, pH 6.5, 298 K.