Abstract
31P NMR relaxation studies from 0.005 to 11.7 T are used to monitor water-soluble inositol 1,2-(cyclic)-phosphate (cIP) binding to phosphatidylinositol-specific phospholipase C spin-labeled at H82C, a position near the active site of the enzyme, and to determine how activating phosphatidylcholine (PC) molecules affect this interaction. We show that, in the absence of an interface, cIP binding to the protein is not rate-limiting, and that lower activation by PC vesicles as opposed to micelles is likely due to hindered product release. The methodology is general and could be used for determining distances in other weakly binding small molecule ligand/protein interactions.
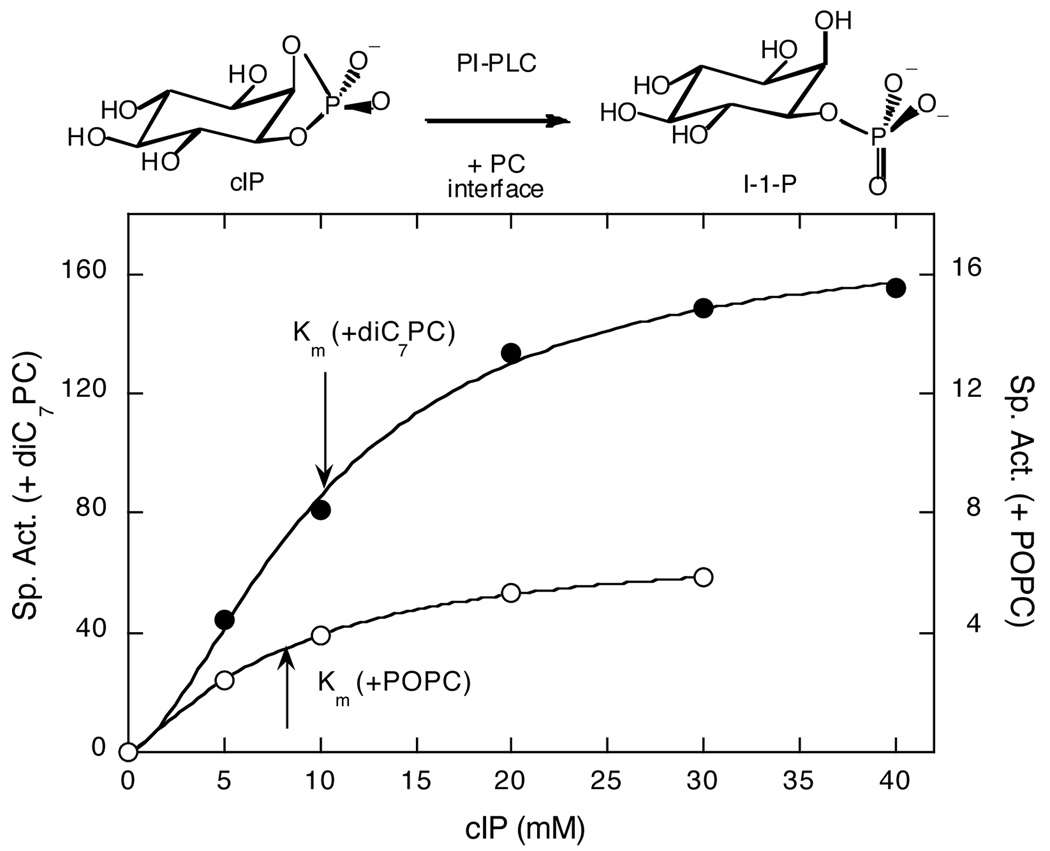
Phosphatidylinositol-specific phospholipase C (PI-PLC) enzymes catalyze the hydrolysis of phosphoinositides in two steps: an initial phosphotransferase reaction to produce diacylglycerol and inositol 1,2-(cyclic)-phosphate (cIP), followed by hydrolysis of the cIP to inositol-1-phosphate. For PI-PLC from Bacillus thuringiensis, water-soluble cIP hydrolysis is slow with a Km >50 mM (1,2), but the specific activity is significantly enhanced by the addition of phosphatidylcholine (PC) interfaces, either micelles such as diheptanoyl-PC (diC7PC) or vesicles of 1-palmitoyl-2-oleoyl-PC (POPC) (2,3). For example, at 5 mM cIP, the addition of diC7PC leads to a 40–50-fold increase in specific activity (4); POPC SUVs added yield a 3–4-fold increase. While both PC interfaces decrease Km, the Vmax is dramatically different (Fig. 1).
Figure 1.
Dependence of PI-PLC specific activity, at 25°C, on cIP concentration with 5 mM diC7PC micelles (●) or 5 mM POPC SUVs (❍) added. Note the 10-fold different specific activity scales for cIP with diC7PC micelles (left) or POPC SUVs (right).
These cIP kinetics raise two interesting questions: (i) does the decreased Km for PC-enhanced cIP hydrolysis reflect a tighter binding of substrate to the enzyme, and (ii) why is Vmax lower with a vesicle activator compared to a micelle surface? To address these, we have used an alternative to standard NMR methods (high resolution field cycling 31P NMR spectroscopy or fc-P-NMR) in which the 31P spins are prepared, and their signals detected, at standard high fields, but their relaxation back toward equilibrium occurs at a lower field, as described in the Supporting Information and elsewhere (5–7). Resonances for cIP and PC species are well separated (cIP at 17 ppm and diC7PC at 0 ppm), at the observation field of 11.7 Tesla (T), and spin-lattice relaxation rates (R1 = 1/T1) can be obtained over a wide range of magnetic field strengths (6,7). The field dependence of R1, from 0.005 up to 11.7 T, can then be analyzed with standard theory (6,7) to obtain correlation times.
For small phosphorus-containing molecules free in solution, the dipolar contribution to 31P relaxation is small, and the 31P R1 is dominated by chemical shift anisotropy (CSA) over most of the accessible field range. But if the small molecule spends part of its time ligated moderately weakly to a larger complex such as an enzyme, then its observed R1 is the weighted average of its small free R1 and the much larger R1 that it would have if bound permanently. The effect is field-dependent and depends on the ratio of ligand/protein as well as proximity of the 31P to the nearby proton dipoles, in either the small molecule or the enzyme, that relax it. A way of enhancing the added effect is to further add an electron-spin label to the protein near a suspected binding site. The much larger magnetic dipole of the electron can have a useful effect on the 31P even with a high ligand/protein ratio.
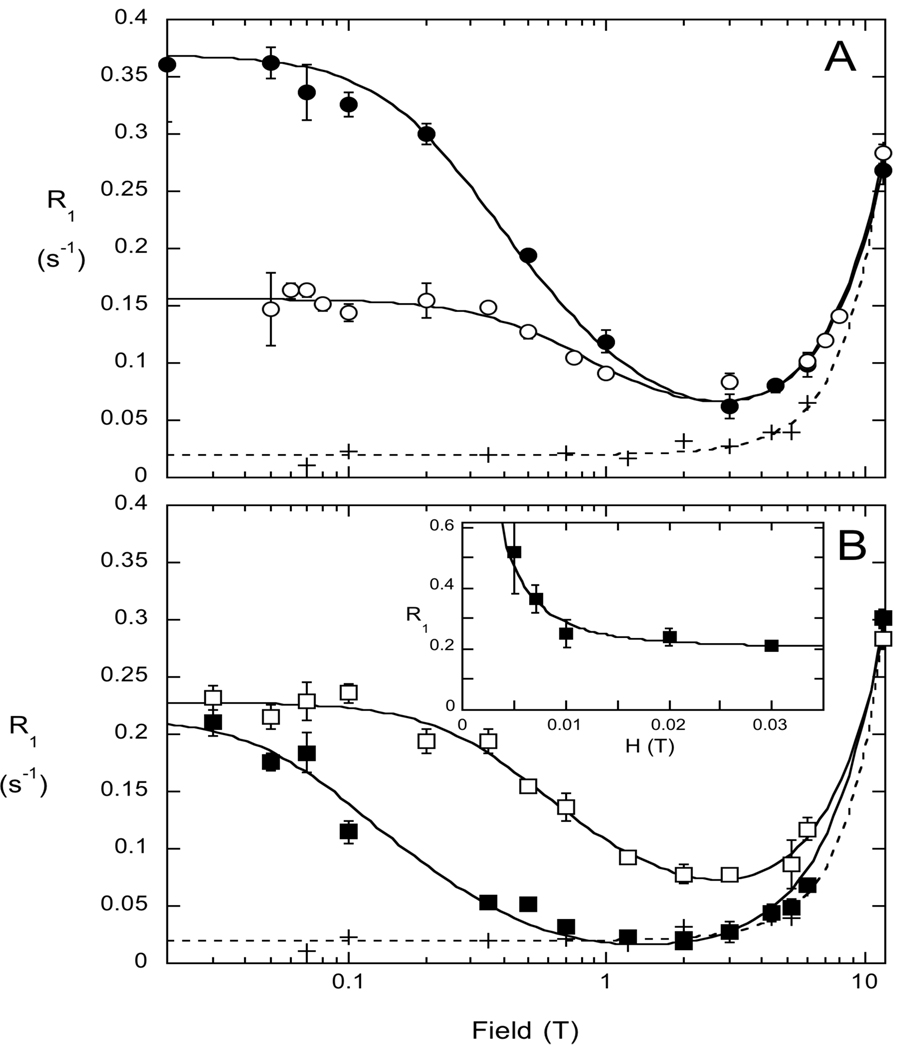
To explore cIP binding to PI-PLC with fc-P-NMR, we used a mutant protein where a key active site residue (His82, the general acid in the proposed mechanism that protonates the DAG anion initially produced in the phosphotransferase step (8)) was altered to cysteine. The H82C enzyme is inactive but should still bind substrate. A nitroxide spin-label (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl-methanethiosulfonate) was attached to this cysteine (H82C-SL) as a way of introducing a much larger dipole in the region where the cIP should bind. The field dependence profiles for cIP (5 mM) with spin-labeled PI-PLC (0.5 mg/ml, 14.4 µM) in the absence and presence of with diC7PC (5 mM) micelles are shown in Fig. 2A. In the absence of protein, cIP exhibits behavior typical for small phosphate esters – the relaxation rate is more or less constant and low below 1 T, with a rise at higher fields due to CSA that follows a square law dependence.
Figure 2.
(A) Field dependence of 5 mM cIP 31P R1 in the absence (+) and presence (❍) of 0.5 mg/ml H82C-SL, as well as with both H82C-SL and 5 mM diC7PC (●) in 50 mM HEPES, pH 7.5, with 1 mM EDTA. (B) Field dependence of the cIP 31P R1 with 5 mM diC6PC (❒) or 5 mM PC SUVs (■) and H82C-SL. For comparison the relaxation profile for cIP in buffer is shown (+). The inset in (B) shows the very low field profile for cIP with the PC SUVs and H82C-SL present.
The presence of 14.4 µM non-spin-labeled H82C (with an excess of DTT present) yielded no significant change in cIP relaxation rates, presumably because the fraction of cIP that binds to enzyme is very small and the 31P-1H interaction in the bound state is relatively small as well. However, when the same amount of spin-labeled PI-PLC (H82C-SL) is added to a sample with cIP alone, there is a substantial increase in R1 below 2 T for cIP (❍, Fig. 2A).
While the binding of cIP to protein is weak, it is specific. In a mixture of three different water-soluble phosphates, cIP, glucose-6-phosphate, and diC4PC, in the presence of H82C-SL (Supplement, Fig. S1), the profiles for the diC4PC and glucose-6-phosphate were identical with or without the H82C-SL indicating they did not bind significantly to the enzyme (or if they did were >20 Å from the site of the spin-label). Only the cIP showed strong relaxation by spin-labeled protein.
The part of the 31P relaxation at each field due to the spin label, ΔR1, was determined by subtraction of the non-spin-label contribution. The residual relaxation data were analyzed using standard relaxation theory (5, 6), modified for a system in fast exchange on the on-enzyme-turnover timescale:
| (1) |
where ΔRP−e(0) is the low-field limit of ΔR1. In eq. 1 we omit the usual terms of theory that involve the electron angular frequency because these are negligibly small owing to the large magnitude of the electron’s frequency compared to ωP. As expected, the τc values so obtained did not depend on total enzyme concentration Eo, and these are presented in Table 1 for the samples shown in Figure 2. In the absence of the diC7PC micelles, the cIP τc increases from 0.019 to 7.1 ns when spin-labeled protein is added. The ΔRP−e(0) of 0.14 s−1 clearly shows that bound cIP must be near the spin-labeled residue. When diC7PC micelles are added, there is a larger effect on ΔRP−e(0), but τc also increases. When POPC SUVs are present, the τc for bound cIP is even longer, 24 ns (■, Fig. 2B), and there is a further increase in R1 below 0.01 T with ~1 µs correlation time (inset, Fig. 2B). This additional low field rise in R1 was not observed for cIP in any of the samples with diC7PC micelles as the activator.
Table 1.
Parameters extracted from fc-P-NMR for cIP (5 mM) with H82C-SL (14.4 µM) in the absence and presence of different PC species (5 mM).
| PC | ΔRP−e (s−1) | τc (ns) |
|---|---|---|
| - | 0.14±0.01 | 7.1±1.0 |
| diC7PC | 0.32±0.01 | 16.8±1.5 |
| diC6PC | 0.21±0.01 | 9.8±1.2 |
| POPC | 0.20±0.02 | 24.4±5.9 |
The values of the parameter ΔRP−e(0) obtained by fitting the data depend on concentrations of total cIP (denoted by [cIP]o) and of total enzyme Eo:
| (2a) |
| (2b) |
Here the term on line (2a) is the fraction of cIP bound to the enzyme relative to the total cIP. In term (2b), S2 is the order parameter of the electron spin-31P dipolar interaction which we take as unity because of the long distance rP−e between the phosphorus of cIP and the electron spin compared to the size of local picosecond motions. We will eventually be interested in the values of rP−e for the different complexes. The last term of (2b) contains standard constants of relaxation theory defined elsewhere (5,6).
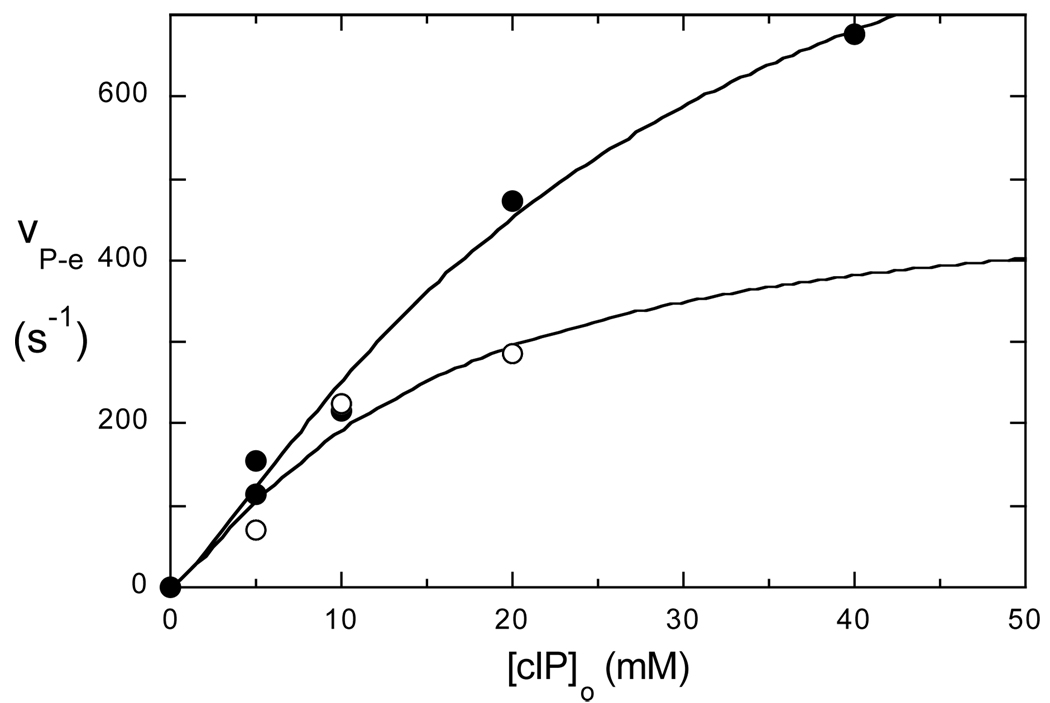
A series of data sets like those of Fig. 2A were obtained for a several total concentrations of enzyme Eo and [cIP]o, the latter chosen so that the R1 of the 31P of cIP would be within the range permitted by the field-cycling apparatus. We assume that binding of cIP to enzyme is a simple bimolecular reaction with binding constant Kd. We define a new parameter vP−e which is equal to ΔRP−e(0) times [cIP]o, divided by total enzyme. The new parameter vP−e is then given by eq. 1 and eq. 2 with the first term (2a) replaced (after one familiar step using mass-action, and the fact that [cIP]o >> Eo) by (1+ [Kd/cIP]0)−1 as shown in eq. 3:
| (3) |
where c is comprised of the terms in (2b). The quantity vP−e is an NMR version of specific activity in enzyme kinetics, and the asymptotic value of vP−e at high concentration of cIP is an NMR analog of an enzymatic Vmax. In Fig. 3, vP−e is plotted versus [cIP]o for the cIP binding to H82C-SL in the presence of either diC7PC micelles or POPC vesicles. The values of Kd for cIP binding to inactive H82C-SL obtained by fitting these curves (25±5 mM with diC7PC and 14±7 mM for POPC SUVs) are roughly twice the Km values obtained from the kinetics (10±1 mM with diC7PC, 8.2±0.3 mM for POPC SUVs). The asymptotic values of vP−e, which are better determined than the Kd values, differ by a factor of 2 with diC7PC micelles leading to more effective relaxation. Dividing the average τc times the constants in (2b) by the asymptote, and taking the sixth root, we get tentative rP−e distances of 7.6±0.1 Å for cIP bound to H82C-SL with diC7PC present and 9.2±0.6 Å when POPC SUVs serve as the activating interface. (They are tentative distances because, in this case, we do not know exactly what motion of the spin-label and 31P that τc represents.) The two distances are significantly different when assessing the experimental error in the vP−e asymptote in Fig. 3, and, more importantly the difference in τc values for the two systems. If we dock cIP into the myo-inositol position in the B. cereus PI-PLC crystal structure (1PTG (10)), and replace His82 with a Cys to which the flexible spin label is attached, the 31P-electron distance is predicted to be in the 5–9 Å, depending on the orientation of the nitroxide group, consistent with the fc-P-NMR analysis.
Figure 3.
Variation of vP−e with total cIP concentration with diC7PC (●) or POPC SUVs (❍) present.
So what do these results imply about the low activity of PI-PLC hydrolyzing cIP without a PC interface? τc/ΔRP−e(0) is proportional to r6 multiplied by the fraction of cIP bound to the enzyme. Since τc/ΔRP−e(0) in Table 1 is the same in the absence and presence of diC7PC, the fraction of cIP at the active site must be similar. Therefore, the lower value of Km in the presence of a PC interface is not the result of increased affinity for the cIP. The PC interface likely promotes a conformational change of the PI-PLC•cIP complex critical for hydrolysis. For ‘monomeric’ diC6PC activation of PI-PLC, the increased τc indicates that the cIP bound to H82C-SL with diC6PC present forms a complex. The larger τc would be consistent with a complex where more than a single diC6PC was interacting with the protein to produce a mini-interface.
As to the lower activation by PC SUVs, there are two notable differences in cIP behavior with micelle versus vesicle. (i) The rP−e is longer (by ~1–2 Å) with POPC SUVs, and (ii) cIP bound to the spin-labeled protein/POPC SUV complex exhibits an increased relaxation rate at very low fields that is not detected for the cIP and protein with diC7PC micelles. PI-PLC binds tightly to PC SUVs with Kd in the 0.04–0.06 mM range (11). Displacement of the His82 catalytic residue could cause the poorer cyclophosphohydrolase activity of the enzyme when POPC SUVs serve as the activating interface. Alternatively, the further increase in the relaxation rate for cIP below 0.02 T (Fig. 2B, inset) with a correlation time that resembles SUV tumbling rates (~1 µs (12)) indicates that the cIP off-rate from the POPC•H82C-SL•cIP complex must be less than 106 s−1, an observation that would be consistent with slow release of IP from the POPC•PI-PLC•IP complex. Interestingly, given that the Kd values for cIP binding to H82C-SL with either interface are similar, a shorter off-rate with SUVs as activators would mean that the on-rate of cIP has been reduced – possibly indicating some occlusion of the active site when the protein is anchored to the vesicle.
Finally, this novel methodology holds promise for identifying binding proximities in other situations. For example, a small amount of protein spin-labeled at a particular site added to a mixture of phosphorus-containing ligands should relax only those ligands that bind near the spin-labeled site – a potentially useful way of identifying non-active-site binding ligands.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Lizbeth Hedstrom, Biology Department, Brandeis Univ. for very helpful discussions.
Footnotes
This research was supported by N.S.F. MCB-0517381 (M.F.R.), N.I.H. GM60418 (M.F.R.), and GM077974 (A.G.R)
Supporting Information Available: Protein preparation and spin-labeling, cIP assays, fc-P-NMR details and data reduction. Material is available at http://pubs.acs.org.
REFERENCES
- 1.Volwerk JJ, Shashidhar MS, Kuppe A, Griffith H. Biochemistry. 1990;29:8056–8062. doi: 10.1021/bi00487a010. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C, Wu Y, Roberts MF. Biochemistry. 1997;36:347–355. doi: 10.1021/bi960601w. [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Qian X, Roberts MF. Biochemistry. 1997;36:10089–10097. doi: 10.1021/bi970846o. [DOI] [PubMed] [Google Scholar]
- 4.Guo S, Zhang X, Seaton BA, Roberts MF. Biochemistry. 2008;47:4201–4210. doi: 10.1021/bi702269u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts MF, Cui Q, Turner CJ, Case DA, Redfield AG. Biochemistry. 2004;43:3637–3650. doi: 10.1021/bi035979q. [DOI] [PubMed] [Google Scholar]
- 6.Roberts MF, Redfield AG. J. Am. Chem. Soc. 2004;126:13765–13777. doi: 10.1021/ja046658k. [DOI] [PubMed] [Google Scholar]
- 7.Redfield AG. Magn. Res. Chem. 2003;41:7553–7768. [Google Scholar]
- 8.Hondal RJ, Zhao Z, Kravchuk AV, Liao H, Riddle SR, Yue X, Bruzik KS, Tsai MD. Biochemistry. 1998;37:4568–4580. doi: 10.1021/bi972646i. [DOI] [PubMed] [Google Scholar]
- 9.Bian J, Roberts MF. J. Coll. Int. Sci. 1992;153:420–428. [Google Scholar]
- 10.Heinz DW, Ryan M, Bullock TL, Griffith OH. EMBO J. 1995;14:3855–3863. doi: 10.1002/j.1460-2075.1995.tb00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pu M, Fang X, Gershenson A, Redfield AG, Roberts MF. J. Biol. Chem. 2009;284:16099–16107. doi: 10.1074/jbc.M809600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts MF, Redfield AG. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17066–17071. doi: 10.1073/pnas.0407565101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.