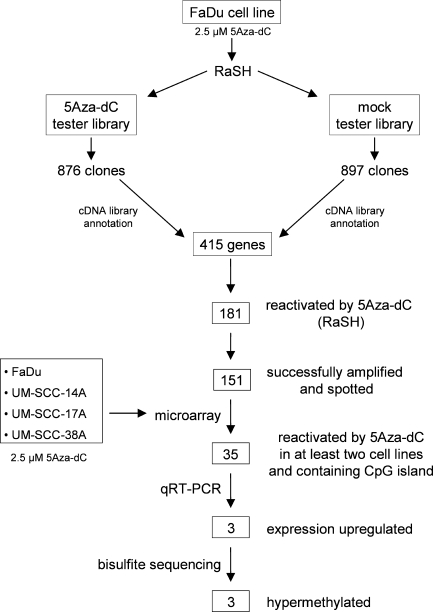
Figure 1.
Flowchart for the identification of differentially methylated genes in HNSCC cell lines. FaDu cell line was treated with 5Aza-dC, and purified mRNA was used to construct RaSH cDNA libraries. A set of 151 nonredundant genes was used to prepare an enriched cDNA platform for microarray analysis. A total of 48 genes were reactivated in at least two cell lines. From them, 35 genes harboring a CpG island located at their 5′ region were submitted to qRT-PCR. Up-regulation of gene expression by 5Aza-dC was confirmed for three genes. Bisulfite sequencing revealed three differentially methylated genes.