Abstract
FimH, the type 1 pilus adhesin of uropathogenic Escherichia coli (UPEC), contains a receptor-binding domain with an acidic binding pocket specific for mannose. The fim operon, and thus type 1 pilus production, is under transcriptional control via phase variation of an invertible promoter element. FimH is critical during urinary tract infection for mediating colonization and invasion of the bladder epithelium and establishment of intracellular bacterial communities (IBCs). In silico analysis of FimH gene sequences from 279 E. coli strains identified specific amino acids evolving under positive selection outside of its mannose-binding pocket. Mutating two of these residues (A27V/V163A) had no effect on phase variation, pilus assembly, or mannose binding in vitro. However, compared to wild-type, this double mutant strain exhibited a 10,000-fold reduction in mouse bladder colonization 24 h after inoculation and was unable to form IBCs even though it bound normally to mannosylated receptors in the urothelium. In contrast, the single A62S mutation altered phase variation, reducing the proportion of piliated cells, reduced mannose binding 8-fold, and decreased bladder colonization 30-fold in vivo compared to wild-type. A phase-locked ON A62S mutant restored virulence to wild-type levels even though in vitro mannose binding remained impaired. Thus, positive selection analysis of FimH has separated mannose binding from in vivo fitness, suggesting that IBC formation is critical for successful infection of the mammalian bladder, providing support for more general use of in silico positive selection analysis to define the molecular underpinnings of bacterial pathogenesis.
Keywords: type 1 pili, uropathogenic Escherichia coli
Virulence factors are genetic determinants of microbial pathogenicity; they increase the fitness of a pathogen during infection and thus should evolve under positive selection (1–3). However, positive selection is not exclusive to virulence factors. Genes may evolve under positive selection due to selective pressures unrelated to virulence, such as those encountered in an environmental reservoir. Nevertheless, the set of genes under positive selection in pathogens should be enriched for virulence factors, and several studies have used positive selection to screen genomic datasets for potential virulence factors (4, 5). In principle, these in silico approaches are attractive for identifying facets of the molecular mechanisms that underlie host-pathogen interactions, as sequence data are easily obtained even in the absence of an experimental model or a genetically manipulable organism. However, direct experimental evidence that positive selection algorithms identify functionally significant mutations in virulence genes is generally lacking (6, 7). To address this issue, we turned to a known virulence factor, FimH, in uropathogenic Escherichia coli (UPEC).
FimH is the mannose-binding adhesin found at the tip of the type 1 pilus produced by most of the Enterobacteriaciae family, including UPEC. Type 1 pili are assembled by the chaperone-usher pathway (8, 9), and FimH plays a key role in initiation of pilus assembly in UPEC (10). Expression of type 1 pili is primarily controlled by transcriptional phase regulation: several recombinase genes mediate inversion of a promoter element controlling transcription of all structural genes required for pilus assembly (11–13). The recombinase genes, in turn, are regulated by several environmental cues (e.g., growth temperature and agitation) (14) and may impact the regulation of other pilus operons (15).
The structural basis for FimH recognition of mannose is known (16). FimH-mediated binding to mannosylated receptors produced by urothelial cells is critical for UPEC to cause bladder infection (cystitis) (17, 18), as it mediates colonization and invasion of superficial umbrella cells lining the luminal surface of the urothelium. UPEC invasion has been reported to involve microtubules (19) as well as several components of lipid rafts such as caveolin-1, an integral membrane protein found in the inner leaflet of the lipid bilayer (20), and Rac1, a member of the Rho family of GTPases (21, 22). After invasion, UPEC can be harbored in exocytic vesicles (23) or escape into the cytoplasm, where they can rapidly replicate into large biofilm-like aggregates (each containing 104–105 bacteria) known as intracellular bacterial communities (IBCs) (24–26).
The IBC pathway has been characterized in a mouse model of UTI (17, 24–26), and IBCs have been documented in humans suffering from UPEC-associated cystitis (27). Furthermore, UPEC strains isolated from the urine of human UTI patients are capable of forming IBCs in multiple mouse backgrounds (28). In mice, IBC formation occurs primarily in superficial umbrella cells (25, 29). Cultured bladder epithelial cells do not generally support IBC formation, presumably because of differences with the terminally differentiated umbrella cells found in vivo. However, biochemical manipulation of cultured human bladder epithelial cells makes them permissive for formation of IBC-like structures (30).
In previous studies, naturally occurring sequence variants in FimH affected fitness in a mouse model of UTI (1, 31), but the corresponding amino acid changes have not been shown to be under positive selection. One amino acid in the signal peptide of FimH has been identified as evolving under positive selection, but this signal peptide is cleaved and not present in the assembled pilus (6).
In this report, we used a maximum likelihood algorithm to conduct a detailed search for positive selection in fimH. Four specific amino acids within the mature protein were found to be evolving under positive selection in a subset of E. coli strains, most of which were cystitis isolates. All of these positively selected amino acid residues (PSAA) reside well outside of the mannose-binding pocket. Mutations in PSAA residues resulted in significant attenuation of UPEC as measured in a mouse model of bladder infection, while mutation of non-PSAA residues had no detectable effect. A mutation in PSAA residue 62 altered type 1 phase regulation, resulting in decreased fitness in vivo. A double mutation in two other PSAA residues did not affect mannose binding in vitro, and the double mutant was able to mediate initial bladder colonization. However, IBC formation was markedly defective, resulting in rapid clearance of UPEC from the bladder. Thus, positive selection has revealed that FimH binding to mannose is necessary but not sufficient for UPEC to cause UTI, and previously unappreciated structure-function relationships in FimH direct an activity downstream of mannose binding that is necessary for IBC formation. We conclude that positive selection is sensitive and specific for identifying amino acids that contribute to in vivo fitness and provides additional insights into virulence factor function.
Results
fimH Gene Is Under Positive Selection in a Subset of UPEC Strains.
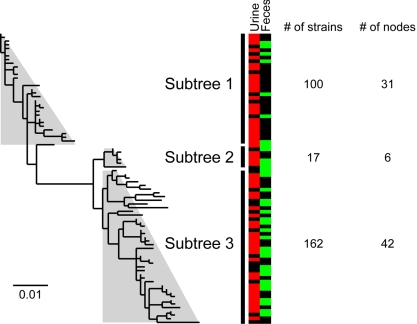
The fimH and fimC genes were sequenced from a collection of 279 E. coli strains including 64 of the 72 ECOR reference strains (32) and 195 clinical isolates obtained from the urine and feces of individuals with acute and recurrent UTI (27) (Table S1). These strains are biased toward cystitis isolates because cystitis is significantly more common than pyelonephritis. FimC, the dedicated chaperone that mediates the assembly of FimH into type 1 pili (33), is not believed to be under positive selection (34, 35) and thus served as a negative control. Maximum likelihood DNA phylograms were inferred using the fimH and fimC sequences from these 279 strains (Fig. 1 and Fig. S1 A and B). Each strain carried one of 79 distinct fimH DNA alleles. Due to synonymous codon usage, these alleles in turn encoded 42 distinct amino acid sequence variants of FimH. To simplify the analysis, phylograms were manually divided into subtrees (see Materials and Methods). Overall, 90% (271/300) of the amino acids in FimH were identical across all sequences (different combinations of mutations in 29 amino acids resulted in the 42 distinct FimH variants alluded to above). Using a maximum likelihood algorithm, we found evidence for positive selection in subtree 1 of fimH but not in any subtree of fimC (Table S2). Residues 27, 62, 66, and 163 of FimH were predicted to be under positive selection in subtree 1. Subtree 1 of fimH was enriched for UPEC strains (P = 0.013, binomial test), depleted for sequences from fecal strains (P < 0.001), and contained the fimH sequences for all UPEC strains for which finished genomes were available at the time of the analysis (strains UTI89, CFT073, and 536).
Fig. 1.
FimH gene phylogeny. Maximum likelihood phylogenetic tree for fimH sequences. Each node represents a unique DNA sequence. Sequence names are omitted for clarity, and light gray trapezoids indicate manually identified subtrees (see Fig. S1 for details). (Scale bar at the bottom left represents mutations per nucleotide.) Color map indicates nodes that include fimH sequences from urine (red) and fecal (green) strains, with black indicating nodes that were found in no urine or fecal strains (some fimH sequences are from strains not isolated from either urine or feces, see Table S1). The number of strains with fimH sequences represented in each subtree is indicated at the right. The number of nodes (distinct DNA sequences) that are represented in each subtree is also indicated at the right (multiple strains can have identical fimH sequences).
Two additional amino acid residues, at positions 70 and 78, were conserved in subtree 1 fimH sequences: 90% (28/31) of subtree 1 sequences encoded Ser-70 and Asn-78, while all fimH sequences in subtrees 2 and 3 encoded Asn-70 and Ser-78. Examination of individual amino acid positions revealed that Asn-70 and Ser-78 are more highly associated with UPEC fimH sequences than any individual PSAA (Fig. 1, color map and Table S3). While Ser-70 and Asn-78 are UPEC-specific amino acid mutations, we found no evidence that they were under positive selection.
Previous studies showed that mutations in PSAA residues 27, 62, and 66 can alter mannose-binding affinity, whereas changes in non-PSAA residues 70 or 78 or PSAA residue 163 had no effect on this activity (31, 36, 37). An A27V mutation in FimH was associated with higher lethality in an i.p. mouse infection model (38), but its impact in the urinary tract has not been directly studied. Besides mannose binding, variation at PSAA residue 66 also affected fitness in a mouse model of UTI and persistence in both the gut and bladder habitats of a single patient (31). None of these residues are located near surfaces known to be important in the protein–protein interactions required for initiation of pilus assembly (39).
Generation of fimH Mutant Strains.
We validated our in silico analysis by testing the effect of mutating PSAA residues and the UPEC-specific, non-PSAA residues 70 and 78. Using the scheme outlined in Fig. S1C, all combinations of mutations in PSAA residues 27, 62, and 163 and non-PSAA 70 and 78 were made in the fimH gene of UTI89, a sequenced human cystitis isolate (5). We also made single mutations in PSAA residue 66. Each amino acid was mutated to its most common variant; for example, all but one of the 79 unique fimH DNA sequences coded for either an Ala or Val at position 27, and fimH in UTI89 encodes Ala-27; therefore, we made an A27V mutation. Thus, we constructed all combinations of A27V, A62S, V163A, and S70N/N78S mutations as well as G66C, G66R, and G66S single mutations in UTI89. A control strain was also made that was subjected to all of the same cloning steps as the mutant strains, but had a wild-type allele of fimH integrated in the last step; this strain is referred to as “wt-control” to differentiate it from the parental UTI89 strain. All of the mutated residues are located at least 20 Å away from the mannose-binding pocket in the 2.8-Å resolution structure of FimH (Fig. S1D).
Mutations in PSAA Affect in Vivo Fitness in the Urinary Tract.
We predicted that mutations in PSAA but not non-PSAA residues would affect in vivo fitness in the urinary tract. We tested this using a C3H/HeN mouse model of UTI. In this inbred strain, infection with UTI89 results in 106–107 colony forming units (CFUs) per bladder 24 h postinfection (hpi). During the first 24 h after a single transurethral inoculation of 1–2 × 107 CFUs, UTI89 binds to and invades the bladder urothelium. This host-pathogen interaction activates several innate defenses (40–42). A fraction of the original inoculum is able to survive or subvert host defenses, invade superficial umbrella cells, escape into the cytoplasm, and rapidly replicate into IBCs. Thus, a single successful invasive bacteria can expand, disperse from the IBC, spread to neighboring urothelial cells, and restart the process of IBC formation (25). IBC formation thus represents an amplification and colonization mechanism for UPEC. Measuring CFUs/bladder at 24 hpi integrates all of these events and thus is a good measure of fitness during UTI.
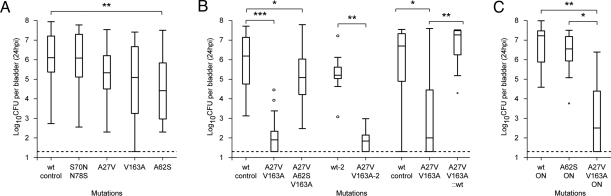
Single mutations in two of the PSAA residues (A27V and V163A) had no significant effect on in vivo fitness in mice (Fig. 2A). However, a double mutation (A27V/V163A) produced a 10,000-fold decrease in CFU/bladder at 24 hpi (P < 0.0001, two-tailed Mann–Whitney test, Fig. 2B). Mutation of PSAA residue 62 (A62S) resulted in a 30-fold reduction in CFU/bladder at 24 hpi compared to the wt-control (P = 0.0006). To rule out possible cloning artifacts, the A27V/V163A mutant and a wild-type control were independently regenerated (yielding strains A27V/V163A-2 and wt-2, respectively). The results were the same as with the original strains (Fig. 2B, middle two lanes). We then reverted the A27V/V163A mutations back to wild type (A27V/V163A::wt); the reverted strain achieved a level of CFUs/bladder at 24 hpi that was indistinguishable from the wt-control strain (Fig. 2B, right three lanes). A triple mutation in all three PSAA residues (A27V/A62S/V163A) had 10-fold lower CFUs/bladder than the wt-control but a 1,000-fold higher level than the A27V/V163A double mutant, indicating that the A62S mutation decreased fitness in the context of the wild-type fimH allele but increased fitness in the context of the A27V/V163A allele (Fig. 2B, third lane). None of the mutants in residue 66 had an effect on fitness in UTI89 (Fig. S2A); however, previous studies have shown a UTI fitness defect due to mutation of residue 66 in a different E. coli strain (with a different wild-type FimH sequence from UTI89) (31). Therefore, mutations in each of the four PSAA significantly alter UPEC fitness in at least one fimH background in a mouse model of UTI.
Fig. 2.
In vivo bladder infection assays at 24 hpi. (A–C) Mutations present are shown on the x axis. A27V/V163A-2 indicates an independently generated clone distinct from the strain labeled A27V/V163A. A27V/V163A::wt denotes a strain with a wild-type fimH gene that was derived from the A27V/V163A mutant strain. The y axis plots the logarithm (base 10) of the bacterial CFU measured in mouse bladders 24 hpi. Dotted line represents the limit of detection. Data are represented as box-and-whisker plots each summarizing data from 20 to 30 mice in (A), 15–30 mice in (B), and 10 mice in (C). “ON” indicates phase locked ON strain. *, P < 0.05, **, P < 0.001, ***, P < 0.0001, two-tailed Mann–Whitney test.
Mutations in Non-PSAA Do Not Affect Fitness.
Mutation of UPEC-specific, non-PSAA residues (S70N/N78S) did not produce significant effects on fitness in mice (Fig. 2A, left two lanes). Because mutation of some PSAA residues only affected fitness in combination with other mutations, we generated the S70N/N78S mutations in combination with A62S and with another naturally occurring allele, A27V/A62S. Interestingly, the A27V/A62S mutant had fitness similar to wt, indicating that the A27V mutation could rescue A62S for the 30-fold decrease in CFU/bladder at 24 hpi. A62S (Fig. 2A, lane 5) and A62S/S70N/N78S (Fig. S2B, lane 2) had similar 30-fold defects compared with wt at 24 hpi and were not significantly different from each other. Similarly, both the A27V/A62S and A27V/A62S/S70N/N78S mutants had similar fitness to wt (Fig. S2B, lanes 3 and 4). Therefore, mutation of UPEC-specific residues 70 and 78 had no effect on in vivo fitness during UTI in mice. Thus, positive selection is both sensitive (PSAA residues have an effect) and specific (non-PSAA have no effect) for in vivo fitness in the bladder habitat.
In Vivo Fitness Correlates with Ability to Form IBCs.
Because CFUs/bladder at 24 hpi encompasses initial colonization as well as subsequent intracellular events, we examined binding and invasion and IBC formation at earlier time points to better define the in vivo fitness defects of the A62S and A27V/V163A mutants. Previous studies of UTI89 have shown that at 1 hpi, bacteria are bound to the surface of, or have invaded into, superficial umbrella cells. Bacteria are also found suspended in the urine at 1 hpi and at all other time points through 24 hpi. After invasion, bacteria can be expelled from the bladder epithelial cell (23) or escape into the cytoplasm of the umbrella cell where IBC formation commences. Replication continues until the entire epithelial cell is occupied by E. coli, which change from a rod to a coccoid shape and divide more slowly. By 6 hpi, a dense, mature IBC has formed (25).
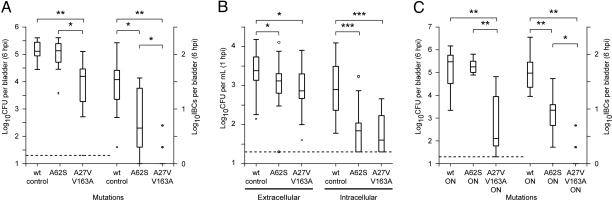
With these observations in mind, we assayed CFUs/bladder and IBC/bladder at 6 hpi (Fig. 3A). A62S had similar CFU/bladder as wt-control, but A27V/V163A was 10-fold reduced. A62S produced 10-fold fewer IBCs than the wt-control, while A27V/V163A formed essentially no IBCs (8/10 mice harbored no detectable IBCs). Thus, differences between A62S and A27V/V163A were apparent by 6 hpi but then dramatically amplified at 24 hpi (compare with Fig. 2 A and B). Because a triple mutant combining all three PSAA mutations (A27V/A62S/V163A) resulted in only mildly decreased CFUs/bladder at 24 hpi (Fig. 2B), we asked whether it could also form IBCs at 6 hpi. The triple mutant was able to rescue the IBC formation defect of the A27V/V163A mutant (P < 0.05, Fig. S2C), although, like the A62S mutant, it did not reach wild-type levels (compare with Fig. 3A).
Fig. 3.
In vivo fitness tests at 1 and 6 hpi. (A) CFU and IBC formation at 6 hpi. Mutations present are shown on the x axis. Left three lanes and y axis plot CFUs/bladder at 6 hpi. Dotted line represents the limit of detection. Right three lanes and y axis show IBCs formed per bladder. (B) In vivo gentamycin protection assay at 1 hpi. Mutations present are shown on the x axis. The y axis indicates the logarithm (base 10) of bacterial CFUs/mL of wash or homogenate. Left three lanes show CFUs per mL wash solution, representing luminal and loosely bound extracellular bacteria in the bladder. Right three lanes are CFU per bladder after treatment with gentamycin and homogenization in 1 mL PBS, and represent intracellular bacteria. (C) Six hours postinfection CFU and IBC formation of phase locked ON strains. Data are shown as in A. *, P < 0.05, **, P < 0.001, ***, P < 0.0001, two-tailed Mann–Whitney test. Each box-and-whisker plot summarizes data for 10 mice.
A gentamycin protection assay (43) was used to differentiate between extracellular and intracellular (invaded) bacteria at 1 hpi (Fig. 3B). Both extracellular and intracellular CFUs of the A62S and A27V/V163A mutants were indistinguishable from each other, but reduced relative to the wt-control (3-fold with respect to extracellular CFUs and 10-fold for intracellular CFUs). We concluded that the severe in vivo fitness defect of the A27V/V163A mutant observed at 6 and 24 hpi is due to processes that occur after invasion of bladder urothelial cells. Moreover, these data further emphasize that IBC formation is a necessary step in UTI in this mouse model (25, 27, 43, 44).
Some UPEC strains do not form IBCs, but this defect is complemented by coinfection with UTI89 (28). Therefore, we evaluated IBC formation by the A62S and A27V/V163A mutants during coinfection with UTI89. Using FimH mutant strains that expressed GFP mixed with an equal number of non-GFP UTI89, we observed no complementation of IBC formation by either mutant at 6 hpi (nearly undetectable GFP-positive IBCs for A27V/V163A; detectable but reduced 10-fold relative to UTI89 for A62S). In addition, there was no dominant-negative effect on UTI89 (i.e., no decrease in GFP-negative IBC formation or CFUs/bladder compared to single infections with UTI89) (data not shown).
In Vivo Fitness Defects Are Specific to the Urinary Tract.
Because positive selection was only found in fimH subtree 1, which was enriched for UPEC strains, we reasoned that selective pressure on PSAA residues A27 and V163 was related to host-pathogen interactions in the bladder. To test the hypothesis that the severe in vivo defect of the A27V/V163A mutant was specific to the urinary tract and not due to generally low fitness in our mouse model, we compared the fitness of the wt-control and A27V/V163A strains (both kanamycin resistant) in competitive mouse gut colonization experiments with UTI89 (kanamycin sensitive). Kanamycin-resistant and -sensitive bacterial CFU/gram of feces was monitored every 3–4 days for 2 weeks, at which point the total CFUs per gram of feces had reached a steady-state level of 1010 (Fig. S2D). As seen in Fig. S2E, the wt-control strain slightly outcompeted UTI89 by 3-fold. The A27V/V163A mutant was indistinguishable from UTI89. These results are in stark contrast to the 10,000-fold difference in bacterial titers seen in the bladder between the wt-control and the A27V/V163A mutant strains, demonstrating that the in vivo fitness defect of the A27V/V163A mutant strain was body habitat-specific and not simply due to use of a mouse model.
In Vitro Mannose Binding Does Not Explain in Vivo Fitness Defects.
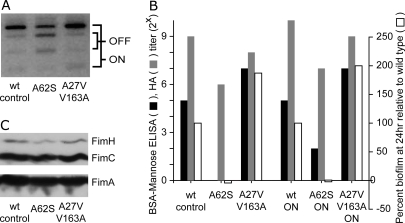
The effect of mutations in the PSAA residues on in vitro phenotypes was also investigated to examine the correlation between (i) fim transcriptional regulation, FimH protein production, mannose binding, biofilm production, plus binding and invasion of cultured 5637 bladder epithelial cells and (ii) the in vivo phenotypes observed in the mouse UTI model (Fig. 4 and Figs. S3 and S4). As noted in the Introduction, type 1 pili expression is under the control of an invertible promoter resulting in phase OFF and phase ON cells, depending on the orientation of the promoter; this process can be monitored by a PCR assay (45). Compared to the wt-control, A62S had a higher proportion of cells in the phase OFF orientation after growth in LB broth without shaking [static culture induces expression of type 1 pili (46)] (Fig. 4A). Accordingly, the A62S strain had (i) consistently lower steady state levels of FimH protein as determined by Western blot analysis of whole cell extracts (Fig. 4B), and (ii) a smaller fraction of piliated cells as determined by electron microscopy (data not shown). No morphological differences in pilus shape or length were seen by electron microscopy. Compared to wt-control, A62S also had 4- to 8-fold lower mannose-inhibitable hemagglutination (HA) titers (n = 10 independent experiments, P < 0.001), 4- to 32-fold reduced binding to BSA conjugated with mannose (n = 2 independent experiments, Fig. 4C and Fig. S3), no detectable ability to form biofilms (n = 2 experiments, Fig. 4C and Fig. S4A), and 10-fold lower binding and invasion of 5637 bladder cells (n = 3 experiments, Fig. S4B). In contrast, the A27V/V163A mutant had no statistically significant defects compared to wt-control in any of these assays. Thus, in vitro, the A62S mutant had impaired FimH function compared with the A27V/V163A mutant. This was surprising given that A62S was much more fit in vivo than A27V/V163A. However, these in vitro HA and binding experiments did not differentiate whether the defects were due to reduced expression or to impaired FimH function. Decreased in vitro HA and binding of the A62S mutant could be due to a higher proportion of bacteria in the phase OFF state compared with the wt-control and A27V/V163A.
Fig. 4.
In vitro characterization of mutant FimH strains. (A) Type 1 pilus phase assay. Mutations are shown at the bottom. The phase orientation in a population of cells grown under type 1 inducing conditions is shown. Bands corresponding to the ON and OFF orientation are indicated. (B) Immunoblot for fimbrial proteins. Bands corresponding to FimA, FimC, and FimH are indicated. (C) In vitro binding assays and in vitro biofilm formation assay. Mutations present are shown on the x axis. “ON” indicates the phase locked ON strain. Binding titers (log base 2) to immobilized BSA-mannose (black bars) and hemagglutination (HA) titers (gray bars) are shown on the left y axis. Relative biofilm formation (white bars) at 24 h for the mutants is plotted on the right y axis, and has been arbitrarily normalized to 100% for the wt-control or wt-ON strains. Data from a single experiment are shown.
Controlling Phase Regulation Rescues the in Vivo Fitness Defect of A62S but Not the Mannose-Binding Defect.
We generated phase locked ON mutants to eliminate FimH expression as a variable by deleting the fimB and fimE recombinase genes that mediate in vitro phase variation in each mutant and isolating the corresponding phase locked mutants. Using phase-locked ON strains, any remaining phenotypic differences should be directly attributable to the effects of PSAA mutations on FimH function.
Using phase locked ON mutants, no phase OFF bacteria were detectable by PCR (data not shown). The relative deficiencies in HA, mannose binding, and binding and invasion of cultured 5637 bladder cells noted in A62S compared to wt-control cells were also observed in the corresponding phase locked ON mutants (denoted A62S-ON and wt-ON). The wt-ON and A27V/V163A-ON continued to behave equivalently in all of the in vitro assays, consistent with results obtained above (Fig. 4C and data not shown). We concluded that the in vitro phenotypes seen were due to an effect of the mutations on FimH function itself and not to altered type 1 pilus regulation.
The phase locked ON mutants were also tested in our mouse model of UTI. Compared to a phase locked wt-ON control strain, the A27V/V163A-ON mutant remained defective in the levels of CFUs/bladder and IBCs/bladder achieved at 6 hpi (Fig. 3C) and 24 hpi (Fig. 2C). The A62S-ON strain still had decreased IBC formation at 6 hpi compared to wt-ON (Fig. 3C). However, CFUs/bladder at both 6 and 24 hpi were indistinguishable from wt-ON (Figs. 2C and 3C); the CFUs/bladder phenotypes of A62S were rescued in the phase locked ON mutant. Thus, the dichotomy between mannose binding and in vivo fitness seen in the A62S and A27V/V163A mutants was not altered by constitutive expression of type 1 pili.
Discussion
The FimH adhesin is required for UPEC to cause UTI in numerous mouse models of infection (17, 18, 26). FimH mediates binding to mannosylated uroplakin proteins present on the luminal surfaces of mouse and human bladder urothelial cells (47). This binding is thought to be the primary molecular feature by which FimH supports UTI (1). Identified amino acid variation in FimH is restricted to 29 residues distributed throughout the protein structure, but the 8 amino acids making up its mannose-binding pocket (16) are invariant in all 188 UPEC strains we studied. Mutation of residues in the mannose binding pocket, such as Q133K, completely abolishes mannose binding activity (16). Accordingly, a Q133K mutant is also severely attenuated in all in vivo assays (Fig. S5). In addition, UPEC strains tend to bind mannose more avidly than fecal E. coli isolates (37), and a fimH allele encoding a protein that binds to mannose more avidly has higher fitness in a mouse model of UTI compared to an allele producing a FimH variant with weaker mannose binding activity (1, 31). Mannose binding is increased by shear forces caused by urine flow, facilitating colonization in the urinary tract (36).
UPEC pathogenesis also involves intracellular infection. FimH is required for invasion of superficial bladder epithelial cells (20, 22). After invasion, epithlelial cells are capable of expelling UPEC, possibly as part of an innate defense (23). However, access to the cytoplasm of superficial umbrella cells provides UPEC with a favorable environment in which they rapidly replicate and aggregate into IBCs (25). These intracellular steps are also dependent on FimH (17). Using an in silico analysis of positive selection and a mouse model of UTI, we have now identified additional residues, outside of the mannose-binding pocket, that are not needed for extracellular mannose binding but are important for intracellular FimH function in vivo. Thus, mutations that did not affect mannose binding but reduced or abolished IBC formation were severely attenuated. This argues that IBC formation is critical in UPEC virulence and serves, in part, as a mechanism for dramatic expansion of bacterial numbers early in infection.
Mutation of residues under positive selection separated mannose binding from in vivo fitness in mice (summarized in Table 1). In vitro, an A62S mutant had measurable functional defects in mannose binding. In vivo, the A62S mutant was less fit than a wt-control at 24 hpi as measured by CFU/bladder. Constitutive expression of the fim operon (in a phase locked A62S-ON strain) rescued pilus assembly defects but not mannose binding in vitro. Surprisingly, in vivo, the A62S-ON strain (compared to a wt-ON control strain) rescued the CFUs/bladder phenotype at 24 hpi seen with the A62S mutant. Therefore, the in vitro mannose-binding defect of A62S is a result of altered FimH function (36, 48), while the in vivo fitness defect of A62S is due to altered fim regulation.
Table 1.
Summary of defects
| Assays | Phase variable strains |
Phase locked ON strains |
|||||
|---|---|---|---|---|---|---|---|
| A62S | A27V/V163A | S70N/N78S | Q133K | A62S-ON | A27V/V163A-ON | ||
| In vitro | Phase regulation, % phase OFF | 50* | 10 | 10 | 50* | 0 | 0 |
| FimH immunoblot | Decreased* | + | + | Decreased* | + | + | |
| Pilus number and morphology | Decreased* | + | + | Decreased* | + | + | |
| HA titer | ↓ 8-fold* | ↓ 2-fold* | + | Undetectable* | ↓ 8-fold* | ↓ 2-fold* | |
| BSA-mannose binding | ↓ 16-fold* | ↑ 4-fold | + | Undetectable* | ↓ 8-fold* | ↑ 4-fold | |
| Biofilm formation | ↓ 128-fold* | ↑ 2-fold | + | ↓ 128-fold* | Undetectable* | ↑ 2-fold | |
| In vivo | Intracellular bacteria at 1 hpi | ↓ 1 log* | ↓ 1 log* | ||||
| IBC formation at 6 hpi | ↓ 1 log* | ↓ >1.5 log* | ↓ >1.5 log* | ↓ 0.5 log* | ↓ >1.5 log* | ||
| CFU/bladder at 24 hpi | ↓ 1.5 log* | ↓ 4 log* | + | ↓ 4 log* | + | ↓ 4 log* | |
All comparisons shown are with wt-control (for phase variable strains on the left) or wt-ON (for phase locked ON strains on the right). Phenotypic defects are marked by *. +, similar to control strain. Blank cells indicate that the experiment was not done.
Despite normal mannose binding in vitro, the A27V/V163A mutant was severely attenuated in vivo. This attenuation was not an artifact of using a mouse model, as there was no severe fitness defect in colonization of the distal gut. None of the in vivo phenotypes of the A27V/V163A mutant were rescued in a phase locked ON strain. Thus, strong mannose binding in the A27V/V163A mutant is not sufficient for in vivo fitness during UTI. The low fitness of A27V/V163A cannot be explained by second site mutations or defects in type 1 pilus assembly or stability; its low fitness is most likely due to a structure-function correlate separate from mannose binding.
We propose that the inability of the A27V/V163A mutant to form IBCs (an intracellular defect) is the reason why it had low in vivo fitness despite strong mannose binding and similar rates of invasion compared to the A62S mutant. Notably, the FimH sequence encoded by the A62S mutant is found in clinical isolates, but the FimH corresponding to the A27V/V163A mutant is not. IBC formation is a general feature of clinical UPEC isolates and is observed in multiple mouse strains (28) and in humans (27). Because IBC formation occurs subsequent to binding and invasion of urothelial cells, the ability to bind mannosylated receptors is a primary and necessary FimH function. Our results suggest that epithelial invasion without IBC formation also leads to bacterial clearance, which may be related to the ability of the bladder epithelial cells to expel UPEC (23). Involvement of FimH in any of several key UPEC infection processes (innate immune evasion, cytoplasmic replication, escaping expulsion, modulation of epithelial apoptosis and exfoliation) could disrupt IBC formation and explain the dichotomy between mannose binding and in vivo fitness seen with the A62S and A27V/V163A mutants.
Of 29 amino acids that vary among FimH sequences from 279 strains, four were identified as being under positive selection. Two additional amino acid positions (70 and 78) are more variable and associated with UPEC strains than any of the PSAA. This association suggests that they may be important for UTI, yet they are not under positive selection and their mutation results in no measurable in vivo effects. In the absence of any phenotypic impact in this and previous studies of polymorphism in residues 70 and 78 (37), it is possible that their association with some UPEC strains is due to nonselective processes such as neutral drift (49) or population subdivision (50). None of the PSAA was located near the conserved mannose-binding pocket of FimH, and examination of the crystal structure of the FimC-FimH-mannose complex revealed no obvious relationship between PSAA mutations and mannose binding. Allosteric effects of FimH mutations have been described and may account for the decreased mannose binding of the A62S mutant (36, 48). However, no structural basis for the severe in vivo fitness defect of the A27V/V163A double mutation is apparent.
Active site and binding pocket residues that are identified by structural analyses are typically a functional subset of conserved amino acids that evolve under negative selection. By identifying a functional subset of variable amino acids, positive selection complements structural and biochemical analyses for probing the function and evolutionary history of a protein. Therefore, at a time when there is an explosive increase in the availability of sequence data for multiple strains of many pathogens, positive selection analysis should be generally useful for exploring the molecular details of myriad host-pathogen interactions.
Materials and Methods
Detailed materials and methods describing positive selection analysis, mutant generation, and in vitro and in vivo assays can be found in the accompanying SI Text.
Supplementary Material
Acknowledgments.
We thank members of the Hultgren and Gordon laboratories for helpful comments, Wandy Beatty for electron microscopy, Sarah Greene and Camille Linton for assistance in creating fimH mutant strains, Bradley Ford for help with crystal structure manipulation, and David O'Donnell and Maria Karlsson for gnotobiotic mouse husbandry. This work was funded by National Institutes of Health Grants DK64540 (S.J.H. and J.I.G.), DK51406 (S.J.H.), AI029549 (S.J.H.), AI48689 (S.J.H.), AI068362 (S.L.C.), and DK081620 (S.L.C.). J.B. was a postdoctoral fellow for the Fonds voor Wetenschappelijk Onderzoek–Vlaanderen (FWO, or Research Foundation–Flanders).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. FJ865583–FJ866110).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902179106/DCSupplemental.
References
- 1.Sokurenko EV, et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiggins FM, Hurst GD, Yang Z. Host-symbiont conflicts: Positive selection on an outer membrane protein of parasitic but not mutualistic Rickettsiaceae. Mol Biol Evol. 2002;19:1341–1349. doi: 10.1093/oxfordjournals.molbev.a004195. [DOI] [PubMed] [Google Scholar]
- 3.Ma W, Guttman DS. Evolution of prokaryotic and eukaryotic virulence effectors. Curr Opin Plant Biol. 2008;11:412–419. doi: 10.1016/j.pbi.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Anisimova M, Bielawski J, Dunn K, Yang Z. Phylogenomic analysis of natural selection pressure in Streptococcus genomes. BMC Evol Biol. 2007;7:154. doi: 10.1186/1471-2148-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen SL, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc Natl Acad Sci USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronald LS, et al. Adaptive mutations in the signal peptide of the type 1 fimbrial adhesin of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2008;105:10937–10942. doi: 10.1073/pnas.0803158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts AJ, Wiedmann M. Allelic exchange and site-directed mutagenesis probe the contribution of ActA amino-acid variability to phosphorylation and virulence-associated phenotypes among Listeria monocytogenes strains. FEMS Microbiol Lett. 2006;254:300–307. doi: 10.1111/j.1574-6968.2005.00041.x. [DOI] [PubMed] [Google Scholar]
- 8.Sauer FG, et al. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 9.Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- 10.Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science. 2008;320:376–379. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]
- 11.Gally DL, Leathart J, Blomfield IC. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 12.Hannan TJ, et al. LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol Microbiol. 2008;67:116–128. doi: 10.1111/j.1365-2958.2007.06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryan A, et al. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect Immun. 2006;74:1072–1083. doi: 10.1128/IAI.74.2.1072-1083.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blomfield IC. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv Microb Physiol. 2001;45:1–49. doi: 10.1016/s0065-2911(01)45001-6. [DOI] [PubMed] [Google Scholar]
- 15.Nowicki B, Rhen M, Vaisanen-Rhen V, Pere A, Korhonen TK. Immunofluorescence study of fimbrial phase variation in Escherichia coli KS71. J Bacteriol. 1984;160:691–695. doi: 10.1128/jb.160.2.691-695.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung CS, et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol. 2002;44:903–915. doi: 10.1046/j.1365-2958.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- 17.Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- 18.Connell I, et al. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhakal BK, Mulvey MA. Uropathogenic Escherichia coli invades host cells via an HDAC6-modulated microtubule-dependent pathway. J Biol Chem. 2009;284:446–454. doi: 10.1074/jbc.M805010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan MJ, Li G, Shin JS, Carson JL, Abraham SN. Bacterial penetration of bladder epithelium through lipid rafts. J Biol Chem. 2004;279:18944–18951. doi: 10.1074/jbc.M400769200. [DOI] [PubMed] [Google Scholar]
- 21.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song J, Bishop BL, Li G, Duncan MJ, Abraham SN. TLR4-initiated and cAMP-mediated abrogation of bacterial invasion of the bladder. Cell Host Microbe. 2007;1:287–298. doi: 10.1016/j.chom.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop BL, et al. Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat Med. 2007;13:625–630. doi: 10.1038/nm1572. [DOI] [PubMed] [Google Scholar]
- 24.Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Justice SS, et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson GG, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 27.Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 2007;4:e329. doi: 10.1371/journal.pmed.0040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garofalo CK, et al. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun. 2007;75:52–60. doi: 10.1128/IAI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mysorekar IU, Hultgren SJ. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc Natl Acad Sci USA. 2006;103:14170–14175. doi: 10.1073/pnas.0602136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berry RE, Klumpp DJ, Schaeffer AJ. Urothelial cultures support intracellular-bacterial community formation by uropathogenic Escherichia coli. Infect Immun. 2009 doi: 10.1128/IAI.00323-09. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissman SJ, et al. Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect Immun. 2007;75:3548–3555. doi: 10.1128/IAI.01963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones CH, et al. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc Natl Acad Sci USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokurenko EV, et al. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol. 2004;21:1373–1383. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- 35.Weissman SJ, et al. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol Microbiol. 2006;59:975–988. doi: 10.1111/j.1365-2958.2005.04985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aprikian P, et al. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J Biol Chem. 2007;282:23437–23446. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- 37.Sokurenko EV, Courtney HS, Maslow J, Siitonen A, Hasty DL. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J Bacteriol. 1995;177:3680–3686. doi: 10.1128/jb.177.13.3680-3686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hommais F, et al. The FimH A27V mutation is pathoadaptive for urovirulence in Escherichia coli B2 phylogenetic group isolates. Infect Immun. 2003;71:3619–3622. doi: 10.1128/IAI.71.6.3619-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munera D, Palomino C, Fernandez LA. Specific residues in the N-terminal domain of FimH stimulate type 1 fimbriae assembly in Escherichia coli following the initial binding of the adhesin to FimD usher. Mol Microbiol. 2008;69:911–925. doi: 10.1111/j.1365-2958.2008.06325.x. [DOI] [PubMed] [Google Scholar]
- 40.Ingersoll MA, Kline KA, Nielsen HV, Hultgren SJ. G-CSF induction early in uropathogenic Escherichia coli infection of the urinary tract modulates host immunity. Cell Microbiol. 2008;10:2568–2578. doi: 10.1111/j.1462-5822.2008.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schilling JD, Martin SM, Hung CS, Lorenz RG, Hultgren SJ. Toll-like receptor 4 on stromal and hematopoietic cells mediates innate resistance to uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4203–4208. doi: 10.1073/pnas.0736473100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schilling JD, et al. CD14- and Toll-like receptor-dependent activation of bladder epithelial cells by lipopolysaccharide and type 1 piliated Escherichia coli. Infect Immun. 2003;71:1470–1480. doi: 10.1128/IAI.71.3.1470-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulvey MA, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 44.Anderson GG, Dodson KW, Hooton TM, Hultgren SJ. Intracellular bacterial communities of uropathogenic Escherichia coli in urinary tract pathogenesis. Trends Microbiol. 2004;12:424–430. doi: 10.1016/j.tim.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Roesch PL, Blomfield IC. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol Microbiol. 1998;27:751–761. doi: 10.1046/j.1365-2958.1998.00720.x. [DOI] [PubMed] [Google Scholar]
- 46.Gally DL, Bogan JA, Eisenstein BI, Blomfield IC. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: Effects of temperature and media. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie B, et al. Distinct glycan structures of uroplakins Ia and Ib: Structural basis for the selective binding of FimH adhesin to uroplakin Ia. J Biol Chem. 2006;281:14644–14653. doi: 10.1074/jbc.M600877200. [DOI] [PubMed] [Google Scholar]
- 48.Pouttu R, et al. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol Microbiol. 1999;31:1747–1757. doi: 10.1046/j.1365-2958.1999.01311.x. [DOI] [PubMed] [Google Scholar]
- 49.Hughes AL. Near neutrality: Leading edge of the neutral theory of molecular evolution. Ann N Y Acad Sci. 2008;1133:162–179. doi: 10.1196/annals.1438.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.da Silva J. Amino acid covariation in a functionally important human immunodeficiency virus type 1 protein region is associated with population subdivision. Genetics. 2009;182:265–275. doi: 10.1534/genetics.108.099853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.