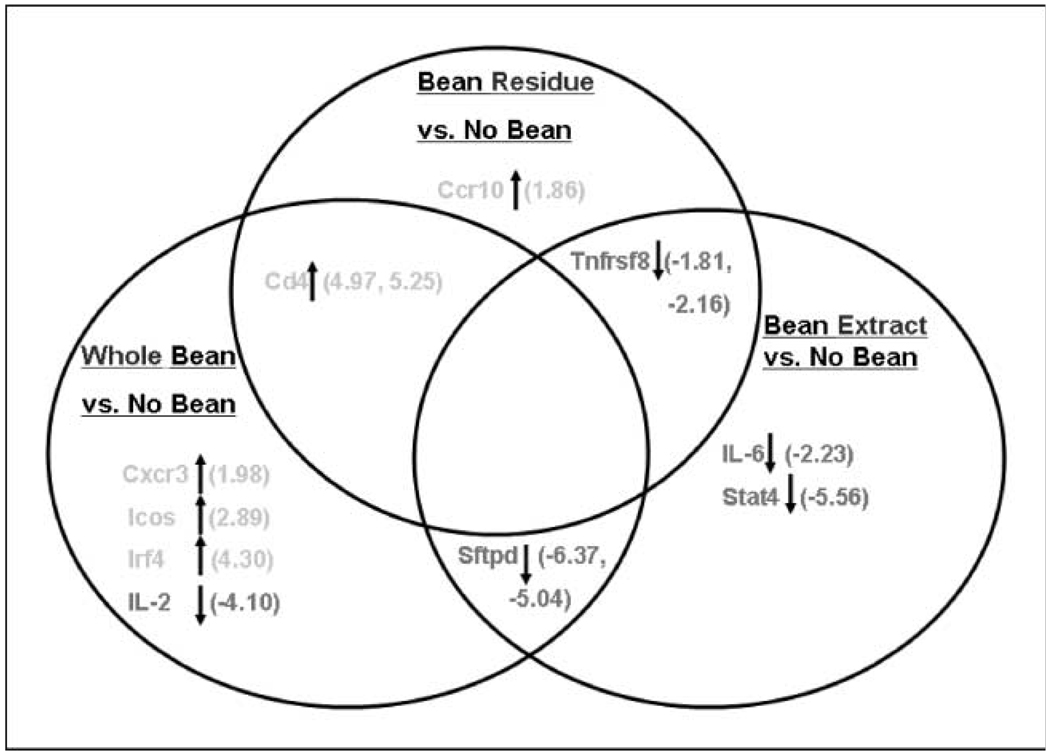
Fig. 3.
Bean diets change gene expression of a subset of inflammation-related cytokines in colon mucosa of AOM-induced ob/ob mice. RNA isolated as described in Materials and Methods. The expression of 84 Th1 Th2 Th3 inflammation-associated genes was analyzed using Superarray real-time quantitative PCR arrays. Shown are Venn diagrams depicting bean-induced gene expression changes that were overlapping or not overlapping between diet groups. Arrows, direction of fold change depicted in parentheses. Criteria for gene selection were P < 0.05 for the difference between bean fed and control, and fold change −1.7. Fold change range for all genes was −2.0 to +6.0. n = 6 mice per group. Analysis was done on non–tumor-bearing mice. Comparison groups included whole bean versus no bean, bean residue versus no bean, and bean extract versus no bean, respectively. Statistical significance: Tnfrsf8 (Bean Extract; P = 0.005) and Stat4 (Bean Extract; P = 0.007). All other genes in the figure had a significant trend where P values ranged from P = 0.012 (Tnfrsf8; Bean Residue) to P = 0.047 (Icos). Genes include CD4 antigen (Cd4), surfactant-associated protein D (Sftpd), TNF receptor superfamily member 8 (Tnfrsf8), chemokine C-X-C motif receptor 3 (Cxcr3), inducible T-cell costimulator (Icos), IFN regulatory factor 4 (Irf4), IL-2, chemokine C-C motif receptor 10 (Ccr10), IL-6, and signal transducer and activator of transcription 4 (Stat4).