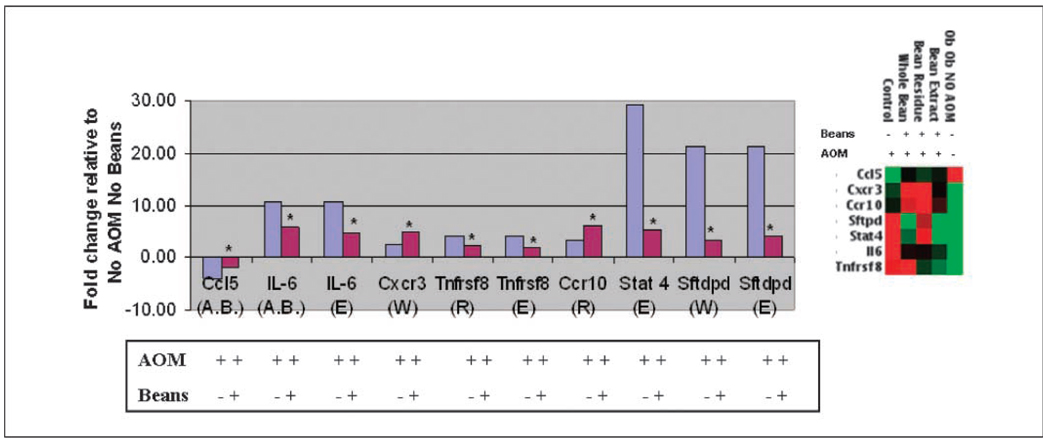
Fig. 5.
Bean diet attenuates some of the gene expression changes induced by AOM in ob/ob mice. Graph represents fold changes relative to non–AOM induced (i.e., −AOM) ob/ob mice, which was the overall comparison group for this analysis. Right, clustergram of fold changes depicted in graph (red, high-expressing genes; green, low-expressing genes). All mice from The Jackson Laboratory (No AOM) were free of colon lesions and tumors, showing normal colon on histopathologic examination. As expected in ob/ob mice, some fat infiltration was observed. In contrast, the AOM-treated mice included some with preneoplastic lesions or tumors as indicated in Fig. 1 (4). Blue columns, ratio of +AOM to −AOM in the absence of beans. Red columns, ratio of +beans to −beans following AOM exposure. The determined (blue columns) and calculated (red columns) fold change values have been ascertained as follows: To identify genes whose expression is induced by AOM (A) and attenuated/counteracted by the bean diets (or repressed by AOM and attenuated by the bean diets) (B), all expression levels were normalized to the level found without AOM or bean diet. The following calculations were done using Superarray results. Expression values were measured on Superarray Th1 Th2 Th3 array plates with one sample run per plate. Calculations were determined using the following x, y, and z groups: x = AOM (+) Beans (−), y = AOM (+) Beans (+), and z = AOM(−) Beans (−). Two ratios were determined: (a) y/x or AOM (+) Beans (+)/AOM (+) Beans (−) and (b) x/z or AOM (+) Beans (−)/AOM (−) Beans (−). To calculate the ratio of expression with AOM with beans to expression without AOM without beans (y/z), the following equation was used: y/z = y/x × x/z. Letter(s) in parentheses following the gene indicates the bean group: A.B., all bean; W, whole bean; R, bean residue; E, bean extract. Total mice per group: No AOM No Bean, n = 5; ob/ob mice, +AOM No Bean, n = 6; +AOM All Bean, n = 18; +AOM whole bean, n = 6; +AOM residue, n = 6; +AOM extract, n = 6. Values for AOM + beans (red columns) were significantly different or had a statistical trend from those for AOM alone (blue columns). Stat4 and Tnfrsf8 genes were significant at the P < 0.001 level. *, genes statistically significant or having a statistical trend. Genes include chemokine C-C motif ligand 5 (Ccl5), IL-6, chemokine C-X-C motif receptor 3 (Cxcr3), TNF receptor superfamily member 8 (Tnfrsf8), chemokine C-C motif receptor 10 (Ccr10), signal transducer and activator of transcription 4 (Stat4), and surfactant associated protein D (Sftpd).