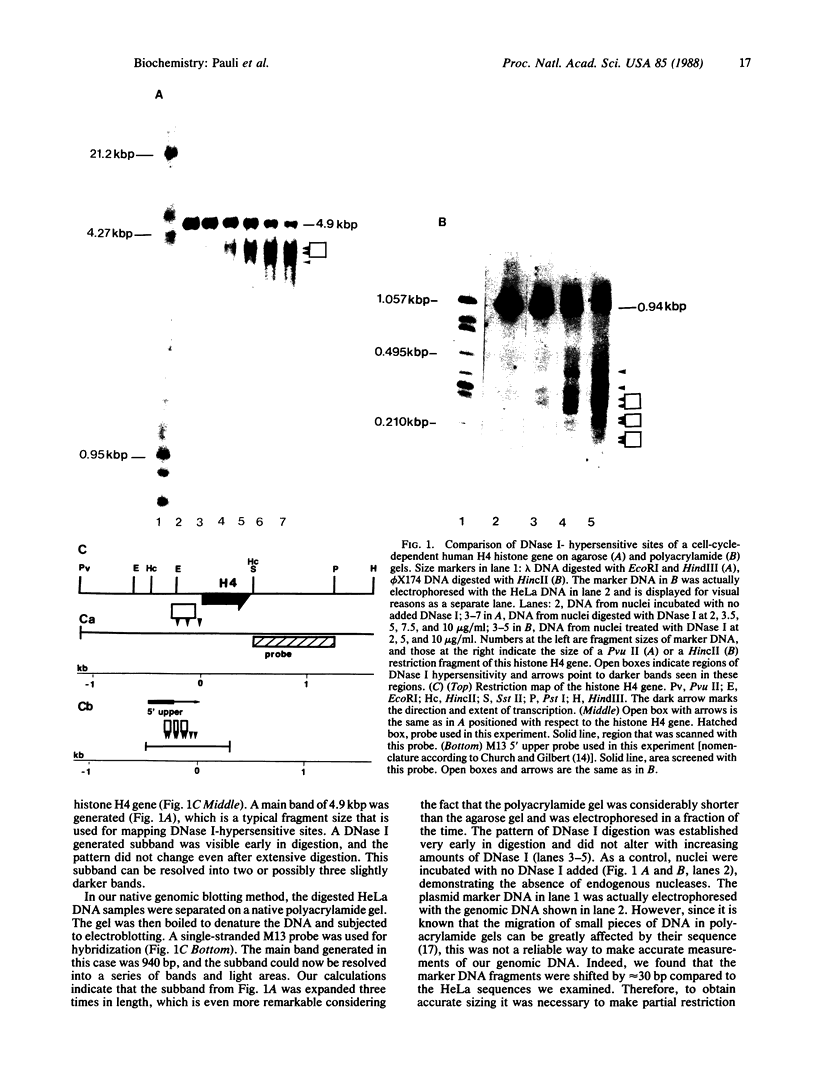
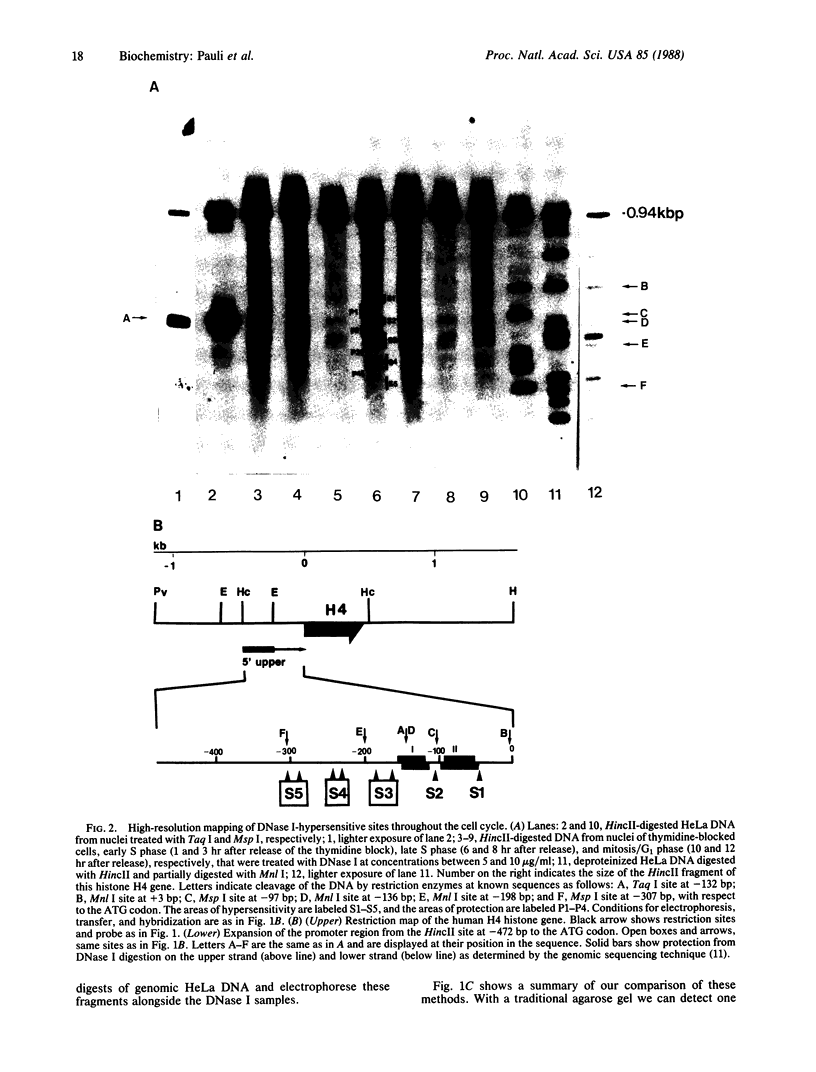
Abstract
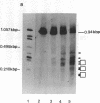
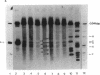
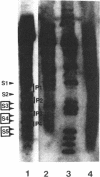
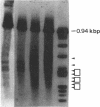
DNase I-hypersensitive sites are observed in the promoter regions of actively expressed genes, potentially active genes, and genes that were once active. We have developed an approach that greatly increases the resolution for mapping these sites by electrophoresing genomic DNA on native polyacrylamide gels prior to electroblotting and hybridization. This improved method has been used to scan the promoter and coding region of a cell-cycle-dependent human histone H4 gene with an accuracy of +/-5-10 base pairs. Protein-DNA interactions can be seen in the autoradiograph as light areas and DNase I-hypersensitive sites as dark bands. Therefore, this method provides a rapid and relatively simple means to accurately localize protein-DNA interactions as well as DNase I-hypersensitive sites, thus directly displaying DNase I hypersensitivity and protein-DNA complexes on one autoradiograph. It also potentially allows the analysis of small changes in DNase I-hypersensitive sites under various biological conditions. With this technique rather large regions of DNA can be screened to determine areas that should be analyzed by more sophisticated methods, such as genomic sequencing or gel retardation assays.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chrysogelos S., Riley D. E., Stein G., Stein J. A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7535–7539. doi: 10.1073/pnas.82.22.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Garrard W. T., Bagi G., Wilson R. F., Bonner J. Partial purification of the template-active fraction of chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2193–2197. doi: 10.1073/pnas.71.6.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- McCutchan T. F., Hansen J. L., Dame J. B., Mullins J. A. Mung bean nuclease cleaves Plasmodium genomic DNA at sites before and after genes. Science. 1984 Aug 10;225(4662):625–628. doi: 10.1126/science.6330899. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Pauli U., Chrysogelos S., Stein G., Stein J., Nick H. Protein-DNA interactions in vivo upstream of a cell cycle-regulated human H4 histone gene. Science. 1987 Jun 5;236(4806):1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- Plumb M., Stein J., Stein G. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucleic Acids Res. 1983 Apr 25;11(8):2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stein G. S., Borun T. W. The synthesis of acidic chromosomal proteins during the cell cycle of HeLa S-3 cells. I. The accelerated accumulation of acidic residual nuclear protein before the initiation of DNA replication. J Cell Biol. 1972 Feb;52(2):292–307. doi: 10.1083/jcb.52.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]