Abstract
In plants, iron homeostasis is tightly regulated to supply sufficient amounts of this metal for an optimal growth while preventing excess accumulation to avoid oxidative stress. To identify new regulators of iron homeostasis, a luciferase-based genetic screen using the Arabidopsis AtFer1 ferritin promoter as a target was developed. This screen identified TIME FOR COFFEE (TIC) as a regulator of AtFer1 gene expression. TIC was previously described as a nuclear regulator of the circadian clock. Mutants in the TIC gene exhibited a chlorotic phenotype rescued by exogenous iron addition and are hypersensitive to iron during the early stages of development. We showed that iron overload-responsive genes are regulated by TIC and by the central oscillator of the circadian clock. TIC represses their expression under low iron conditions, and its activity requires light and light/dark cycles. Regarding AtFer1, this repression is independent of the previously characterized cis-acting element iron-dependent regulatory sequence, known to be involved in AtFer1 repression. These results showed that the regulation of iron homeostasis in plants is a major output of the TIC- and central oscillator-dependent signaling pathways.
Introduction
Iron is an essential element for all organisms. However, its physiological concentration needs to be tightly regulated because of its high reactivity with oxygen (1). The regulatory mechanisms controlling iron homeostasis in bacteria and animals are well documented (2, 3), in contrast with the lack of knowledge in plants.
This last decade, a wealth of information has been obtained on the molecular characterization of genes involved in iron acquisition by roots, in iron allocation to various organs, and in subcellular compartmentalization of iron in plants (reviewed in Refs. 4 and 5). The present challenge in this field concerns our understanding of the integration of these functions at the whole plant level and the signaling networks and regulatory molecules responsible for the control of iron dynamics in plants. Although the iron regulation of genes involved in uptake and storage has been widely studied, only a few components of the iron signaling pathway are known. Furthermore, studies regarding this aspect have, up until now, focused on the identification of regulatory proteins that function in the iron deficiency signaling pathway (reviewed in Refs. 6 and 7). Hence, genes involved in iron excess signaling pathways have yet to be identified in plants. One of the primary responses to iron excess is a quick accumulation of a large amount of ferritins in plastids (reviewed in Ref. 6). Ferritins play an essential role in iron homeostasis by sequestering iron in a bio-available and nontoxic form, thus preventing oxidative stress (8). Among the four ferritin genes of Arabidopsis thaliana, AtFer1 is the most highly expressed in response to iron excess (8), making it an ideal model for studying iron homeostasis regulation. Furthermore, AtFer1-transcriptional regulation by the iron status occurs through a repression/derepression mechanism. Experiments based on serial deletions and site-directed mutagenesis of the AtFer1 ferritin promoter region allowed the identification of a 15-bp cis-acting element necessary for the iron-dependent regulation of the AtFer1 transcription (9). This sequence, named iron-dependent regulatory sequence (IDRS),3 is involved in the repression of AtFer1 expression under iron deficiency (9, 10). Thus, iron addition leads to the derepression of AtFer1 rather than to its induction. To identify components of the iron signaling pathway, we screened for mutants impaired in the AtFer1 repression/derepression in response to iron status. We used a pAtFer1::LUC reporter line to identify ethyl methanesulfonate (11) mutants in Arabidopsis harboring a high LUC activity under repressive conditions, i.e. low iron. Two mutants, exhibiting a high LUC activity and a 10-fold increase in AtFer1 mRNA accumulation compared with wild type plants, were isolated and named dif3 and dif6 (deregulated in ferritin). They were allelic and mutated in the TIC (Time for Coffee) gene, which encodes a nuclear regulator of the circadian clock (12, 13). We showed that the mutation in TIC led to the derepression of AtFer1 under repressive conditions. The TIC-mediated AtFer1 regulation was circadian clock-dependent and did not occur through the cis-element IDRS. Our results showed that TIC and a functional central oscillator of the circadian clock are required for the expression of iron-regulated genes. This work points out that when plants do not experience iron excess, iron homeostasis is regulated through a circadian clock-dependent pathway, leading to the modulation of iron-responsive genes.
EXPERIMENTAL PROCEDURES
Plant Material
The wild type A. thaliana ecotypes used were Columbia-0 (Col) and Wassilevskija (Ws). The mutants dif3, dif6, and tic-2 were in the Col genetic background; tic-1, elf4-1, cca1-11, and lhy-21 were in the Ws genetic background.
Growth Conditions
For hydroponic cultures, the plants were grown for 6 weeks in a growth chamber (250 μmol·m−2·s−1, relative humidity 70%, 8 h of light at 23 °C/16 h of dark at 20 °C). The seeds were sown on wet sand laid on a polystyrene raft floating on water. After 2 weeks on water, the rafts were transferred to a nutrient solution containing the following elements: 0.5 mm KNO3, 0.25 mm Ca(NO3)2, 1 mm MgSO4, 1 mm KH2PO4, 100 μm Fe(III)-Na-EDTA, 50 μm H3BO3, 19 μm MnCl2, 1 μm CuSO4, 10 μm ZnCl2, 0.02 μm MoO4Na2, pH 5.8. The nutrient solutions were renewed every week. The treatments were performed on 6-week-old plants. For iron deficiency treatment, the roots were washed twice with 0.3 mm bathophenanthroline disulfonic acid and 5.7 mm Na2S2O4 to remove apoplasmic iron. Then the rafts were transferred to the initial nutrient solution but without iron added. Iron overload treatment was performed after 1 week of culture in the iron-deficient medium, by adding 500 μm iron citrate for 5 h (14). In a greenhouse, the plants were grown on Humin substrate N2 (Neuhaus, Klasmann-Deilmann, Germany) at 23 °C, with a sunlight intensity limited to 300 μmol·m−2·s−1 and 16 h of light/8 h of dark.
For in vitro culture, the seeds were surface-sterilized by soaking in a solution containing Bayrochlor 1.5% (w/v) (Indusco France) and ethanol 50% for 30 min under agitation, rinsed three times in ethanol, and dried overnight under a sterile air flow. Sterilized seeds were sown on plates containing a half Murashige and Skoog standard medium (Sigma M0654) supplemented with 100 μm H3BO3, 100 μm MnSO4, 30 μm ZnSO4, 5 μm KI, 1 μm Na2MoO4, 0.11 μm CoCl2, 0.10 μm CuSO4, 0.5 g·liter−1 2-morpholinoethane sulfonic acid (Sigma), 1% (w/v) sucrose (Sigma), 7 g·liter−1 agar, pH 5.8. The plates were placed in a growth chamber (300 μmol·m−2·s−1, relative humidity 70%, 16 h of light at 23 °C/8 h of dark at 20 °C). Ferrozine (3-(2-pyridinyl)-5,6-diphenyl-1,2,4-triazine-4′,4″-disulfonic acid sodium salt) was added at a final concentration of 200 μm for short term iron deficiency. A solution of 500 μm iron citrate was sprayed on plants for short term iron excess experiments.
Circadian Phase Response Analysis
The plantlets were entrained on soil for 2 weeks in 16 h of light/8 h of dark conditions in a growth chamber (20 °C, 250 μmol·m−2·s−1) and then moved into continuous light or kept in 16 h of light/8 h of dark. Triplicate samples were harvested every 4 h for Q-PCR analysis.
Dark-induced Response Analysis
The plantlets were entrained on soil for 2 weeks in 16 h of light/8 h of dark conditions in a growth chamber (20 °C, 250 μmol·m−2·s−1) and then moved into continuous darkness for 3 days. T0 was 30 min before the transfer and the lighting. Each day, the triplicate samples were harvested 30 min before the subjective dawn for Q-PCR analysis. The third day, at the subjective dawn, the plants were put back in the initial growth chamber (20 °C, 250 μmol·m−2·s−1), and triplicate samples were collected 1, 3, and 5 h after transfer.
Mutant Screening and Map-based Cloning
A transgenic line, carrying the promoter of AtFer1 fused to the LUC reporter gene, was used for mutant screening. The NheI/XbaI-digested luc+ fragment, excised from the pSP-luc+ vector (Promega, WI), was inserted into the SpeI/XbaI-digested pBluescript II KS+ (pKS+, Stratagene). Afterward, the SalI/NcoI-digested fragment of the At1400IDRS construct (9) was inserted into the pKS-luc+. The resulting At1400IDRS-luc+ fragment was finally inserted into the KpnI/SacI-digested pMOG402 vector (MOGEN International). The resulting vector was introduced into Agrobacterium tumefasciens GV3101 strain and used to transform A. thaliana using the floral dip method (15). A mono-insertional and homozygous line was selected. Approximately 20,000 M1 seeds were mutagenized with 0.3% EMS for 10 h at 25 °C and sown on soil in a greenhouse. M2 seeds were collected in pools of 200 M1 plants. For the screening, 13-day-old M2 seedlings grown in vitro were sprayed with 1 mm luciferin (Promega) in 0.01% Triton X-100 and assayed for LUC bioluminescence using a CCD camera system (Hamamatsu C4880, Hamamatsu Photonics K.K., Joko, Japon). Bioluminescence images were processed using the software HiPic32 (Hamamatsu Photonics).
The rough mapping was performed thanks to markers described by Montpellier Institut National de la Recherche Agronomique and the Arabidopsis Information Resource. The fine mapping was performed by using tuned markers from the Marker Tracker data base at the University of Toronto. Genomic DNA from dif3 mutant plants was extracted, and the mapped region was amplified by PCR as a set of 1 kbp of DNA fragments, each contiguous fragment overlapping on 150 bp. These DNA fragments were sequenced. The obtained nucleotide sequences were assembled with the SeqMan software (DNASTAR, Lasergene) and then aligned with the Col wild type DNA sequence to determine the mutation positions.
T-DNA Insertion Mapping
SAIL_753_E03 (tic-2) harbors a T-DNA insertion that was localized in the fourth exon of TIC after sequencing of the PCR product using the T-DNA-specific primer LB3A and the TIC-specific primers S31-F and S31-R. The pAtFer1::LUC transgene was mapped to chromosome 4 by thermal asymmetric interlaced PCR (16) in the intergenic region between At4g14820 and At4g14830 (data not shown). The position was confirmed by sequencing with LW-AD2, P1, and P2 primers. The insertion of the At1400m*IDRS:GUS was similarly recovered in the At2g38120 gene with LW-AD2 and P3-R primers. All of the primers used are presented in supplemental Table S1. For the cca1-11 and lhy-21 mutants, primer pairs were designed along the CCA1 and LHY genomic sequence. Each primer pair was combined with the left border primer, specific of the T-DNA (JL202). The CCA1–2F/JL202 and LHY-3R/JL202 produced amplification, and their PCR products were subsequently sequenced to confirm the insertion. The T-DNA location is presented in supplemental Fig. S2.
AtFer1 Overexpressing Line
The coding sequence of the mature AtFER1 protein (17) was digested BamHI/SacI and then inserted into the pSC51 vector. To obtain the pSC51 vector, the EcoRI/BamH1/BamI-digested GUS reporter gene fused to the NOS terminator was excised from the pBI121 (GenBankTM accession number AF485783) and cloned into the pMOG402 vector. The resulting vector was named pMOG-GUSt, and a 35S promoter was inserted into this vector by a SacI/HindII digestion, thus creating the pSC51 vector. The pSC51 vector with the AtFer1 fragment was introduced into A. tumefasciens as described for the pAtFer1::LUC construct. Homozygous and mono-insertional lines were selected, and a line exhibiting a high AtFer1 mRNA accumulation was chosen and named p35S::AtFer1 Col.
RNA Isolation and Q-PCR
RNAs were isolated from rosettes or roots with the TRIzol reagent according to the manufacturer's instructions (Invitrogen). A DNase (Promega) treatment was performed on 5 μg of total RNAs to prevent genomic DNA contamination. RNA samples were subsequently used for reverse transcription (Moloney murine leukemia virus reverse transcriptase; Promega) with anchored oligo(dT)15 (Promega) and dNTPs 0.4 mm. The cDNAs were diluted twice with water, and 1 μl of each cDNA sample was assayed by Q-PCR in a LightCycler (Roche Applied Science) using FastStart DNA Master PLUS Syber Green I (Roche Applied Science). The amplification efficiency was assessed relative to a sample standard. Expression levels were calculated relatively to the appropriate housekeeping gene (HK) using the comparative threshold cycle method, where Ct represented the threshold cycle for target amplification: ΔCt = Ct,gene of interest − Ct,HK and the relative transcript level (RTL) was calculated as follow: RTL = 10 × 2−ΔCt. The primers are presented in supplemental Table S1.
Immunodetection of Ferritins
Five microliters of buffer (50 mm Tris-HCl, 5% (v/v) SDS, 0.72 m β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, 25 mm EDTA, 0.1% (w/v) bathophenanthroline disulfonic acid) were added per mg of plant tissue fresh weight. After 15 min at 4 °C, the samples were centrifuged for 30 min at 13,000 × g. Protein content was measured using bovine serum albumin as a standard (18). The protein samples were resolved by electrophoresis in an SDS-13% polyacrylamide gel (19) and then electroblotted onto polyvinylidene difluoride Hybond-P (Amersham Biosciences). Ferritin immunodetection was performed using a rabbit polyclonal antiserum raised against AtFER1 (17) recognizing the four ferritin subunits (8) and the Immobilon Western blotting kit (Millipore). A loading control was performed on a Coomassie Blue gel (Brilliant Blue R concentrate; Sigma).
LUC Reporter Gene Activity
The samples were grounded, suspended in 300 μl of 100 mm Na2HPO4 and centrifuged at 13,000 × g. Fifty microliters of Steady-Glo luciferase assay system (Promega) was added to 50 μl of protein extract supernatant. LUC activity was measured for 1 s after a 15-min incubation at room temperature and normalized to protein concentration, quantified with Bradford reagent (Bio-Rad) using bovine serum albumin as a standard.
Determination of Photosynthetic Pigment Content
The total pigment content was measured at 644.8, 661.6, and 470 nm after extraction with 2 ml of acetone for 50 mg of ground tissues (20). The pigment content was calculated as follows.
 |
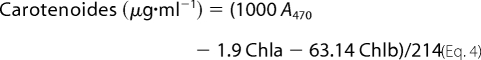 |
Determination of Iron Content
The measurement of total iron content was performed as previously described (21). The samples were mineralized (22), and iron concentration was measured by the absorbance of 1% Fe2+-bathophenanthroline disulfonic acid at 535 nm using thioglycolic acid (Sigma) as a reducing agent. Determination of iron content was performed using a range of a standard iron solution (Carlo Erba).
RESULTS
Identification of A. thaliana Mutants Affected in the Regulation of the AtFer1 Ferritin Gene
To identify genes involved in the transcriptional regulation of AtFer1 in response to iron, transgenic lines in the Col-0 background carrying the AtFer1 promoter fused to the LUC reporter gene were produced for the screen. Because AtFer1 has been shown to be repressed in low iron conditions (9) and derepressed upon iron addition, the first genetic screen we performed aimed at identifying mutants derepressed under low iron conditions. A transgenic line exhibiting the lowest LUC activity under low iron, i.e. the highest repression of the promoter activity, was selected for EMS mutagenesis. The screening was performed on plants grown on a medium with low iron concentration (10 μm iron-EDTA). Candidate mutants harboring a high LUC bioluminescence were transferred on soil to obtain M3 seeds. The LUC phenotype was confirmed in the M3 progeny, and the candidate mutants were validated by selecting those overaccumulating the endogenous AtFer1 mRNAs under low iron conditions. Confirmed mutants were named dif (deregulated in ferritin). Among the eight mutants identified, the mutant line dif3, exhibiting a high LUC expression, was selected for further analysis. This mutant line was backcrossed with a transgenic line. The segregation of the F2-progeny demonstrated that the mutant phenotype is caused by a recessive mutation independent of the reporter gene. For map-based cloning of the mutation, a dif3 plant of the M4 progeny was crossed with Ler. Genomic DNA was prepared from the LUC-positive class (n = 220) of the F2 population (n = 1250) from this out-cross. Using cleaved-amplified polymorphic sequence and simple sequence length polymorphism markers, dif3 was mapped to the upper arm of chromosome 3 at 28 centiMorgan on the Arabidopsis genetic map, between the simple sequence length polymorphism marker CER464947 and the cleaved-amplified polymorphic sequence marker Arlim15.1 (Fig. 1A). Afterward, for fine mapping, additional cleaved-amplified polymorphic sequence markers were used to surround the dif3 mutation into a 136-kbp interval between markers J22310 and J22700, localized respectively in the genes At3g22310 and At3g22700 (Fig. 1A). This genomic DNA fragment was sequenced, and only one mutation was identified, corresponding to a C-T change leading to a premature stop codon in the tenth exon of the TIC gene (At3g22380) (Fig. 1B). Another mutant, called dif6, was isolated from an independent pool of mutagenized seeds and crossed with dif3 for an allelism test. The two mutants were shown to be allelic based on the high LUC activity of plants from the F1 progeny (data not shown), suggesting that the mutations in each line affected the same gene. Therefore, the TIC gene of dif6 was sequenced, and a mutation corresponding to a C-T change was identified in the fourth exon of TIC (Fig. 1B). Two other tic alleles were used to confirm the positional cloning of dif3: the EMS mutant tic-1 in Ws background and the T-DNA insertion mutant tic-2 in Col background (12, 13) (Fig. 1B). Grown on soil, dif3 and dif6 exhibited a 10-fold increase in AtFer1 mRNA abundance compared with transgenic line, tic-2 exhibited a 16-fold increase compared with Col, and tic-1 exhibited a 9-fold increase compared with Ws (Fig. 1C). Hence, the deregulation of the AtFer1 gene expression in four independent tic alleles confirmed the mapping data. The deregulation observed at the mRNA level also occurred at the protein level because a higher abundance of ferritin proteins was observed in dif3 (Fig. 1D) and in tic-2 (Fig. 1E) compared with the wild type plants. Because dif3 carried mutations other than the one in the TIC gene and because similar results were obtained for the EMS mutant dif3 and the T-DNA insertion mutant tic-2, the latter was used for subsequent experiments.
FIGURE 1.
Identification of A. thaliana mutants affected in ferritin regulation. A, map-based cloning of dif3. The marker names are in bold type. The JXXXXX nomenclature indicates that the corresponding marker was designed in the At3gXXXXX gene (Marker Tracker). The number of recombinant lines for each marker over the total number of lines genotyped is in italics. The dif3 locus was mapped between J22310 and J22700, a 136-kbp region that was sequenced. B, structure of the TIC gene. The boxes represent exons and lines introns. In the predicted structure of TIC adapted from Ref. 12, the mutation in dif3 as in dif6 was a C-T change in TIC, tic-2 is a T-DNA insertion mutant, and tic-1 an EMS mutant. C, expression of AtFer1 in four tic alleles grown on soil. Leaves from 3-week-old plants were collected from each line. The relative transcript level of AtFer1 was measured by reverse transcriptase Q-PCR, using the SAND (A2g28390) housekeeping gene (58) as a control. the values are the means ± S.D. (n = 3). D, ferritin accumulation in leaves of dif3. E, ferritin accumulation in leaves of tic-2. D and E, plants were grown as in C. Twenty micrograms of total proteins, extracted from leaves, were loaded per lane, and immunodetection was performed using an anti-FER1 serum (17). The upper panel shows the autoradiography (Ferr.), and the lower panel shows the Coomassie Brilliant Blue staining used as a loading control (Coom.).
Phenotypical Characterization of the tic-2 Mutant
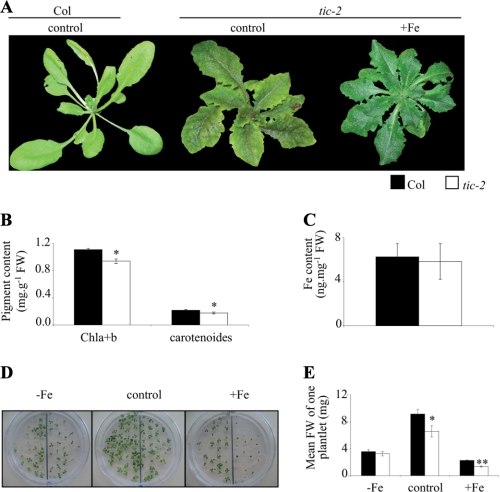
We performed some macroscopic and molecular analysis of the tic-2 mutant to estimate the impact of the mutation on plant growth and development. In a greenhouse, tic-2 exhibited a strong internerval chlorosis compared with the wild type (Fig. 2A). The same chlorosis was observed with the three other mutant alleles (supplemental Fig. S1). This kind of chlorosis was similar to the one observed in the case of iron deficiency (5, 23, 24). When tic-2 plants were irrigated with iron, the chlorotic phenotype was rescued, indicating that it was due to iron deficiency (Fig. 2A). Pigment contents were determined in Col and tic-2 grown in control conditions. The tic-2 mutant had a significant decrease of both chlorophyll and carotenoid contents (18 and 22% decrease, respectively; Fig. 2B). The iron content in leaves of Col and tic-2 was similar (Fig. 2C), indicating that the chlorotic phenotype observed for tic-2 was not due to an iron uptake or translocation from roots to shoots defect. Because the mutant seemed to be sensitive to iron supply on soil, its early development was analyzed in various iron nutrition conditions: deficiency (0 μm iron-EDTA, −Fe), sufficiency (50 μm iron-EDTA, control), and excess (500 μm iron-EDTA, +Fe). Grown in vitro, tic-2 development was affected on both iron-sufficient and iron excess media, but not on iron-deficient medium (Fig. 2D). Except under iron deficiency, tic-2 plantlets were smaller revealing sensitivity to iron. The production of fresh weight of Col and tic-2 plantlets was measured. On control and +Fe media, tic-2 biomass was, respectively, 30 and 40% lower than Col (Fig. 2E). This whole set of results showed that a mutation in the TIC gene led to alterations in iron homeostasis and to sensitivity to iron excess.
FIGURE 2.
Phenotypical analysis of the tic-2 mutant in response to iron status. A, representative 4-week-old Col and tic-2 plants grown on soil in a greenhouse and irrigated with water (control) or irrigated with iron-EDDHA 600 μm (+Fe). B, determination of pigment content in 4-week-old Col and tic-2 plants grown on soil in a greenhouse and irrigated with water. FW, fresh weight. C, determination of iron content in the leaves of 4-week-old Col and tic-2 plants grown on soil in a greenhouse and irrigated with water. Shown are the means ± S.D. of nine measurements (three replicates/experiment performed three times). D, iron sensitivity of plants grown as followed. The plants were grown in vitro during 2 weeks on medium without the addition of iron (−Fe), with the addition of iron-EDTA 50 μm (control), or with the addition of Fe- EDTA 500 μm (+Fe). Col is on the left half of each panel, and tic-2 on the right half. E, mean fresh weight of one plantlet. The plants were grown as described for D. The weight of a plantlet was calculated as followed: weight of a plantlet = (fresh weight of plantlets of a genotype from the same plate)/(number of plantlets of a genotype from the same plate). B and E, shown are the means ± S.D. (n = 3). B, C, and E, *, p < 0.05; **, p < 0.01; black and white bars represent, respectively, Col and tic-2.
TIC-mediated AtFer1 Regulation Is Circadian Clock-dependent
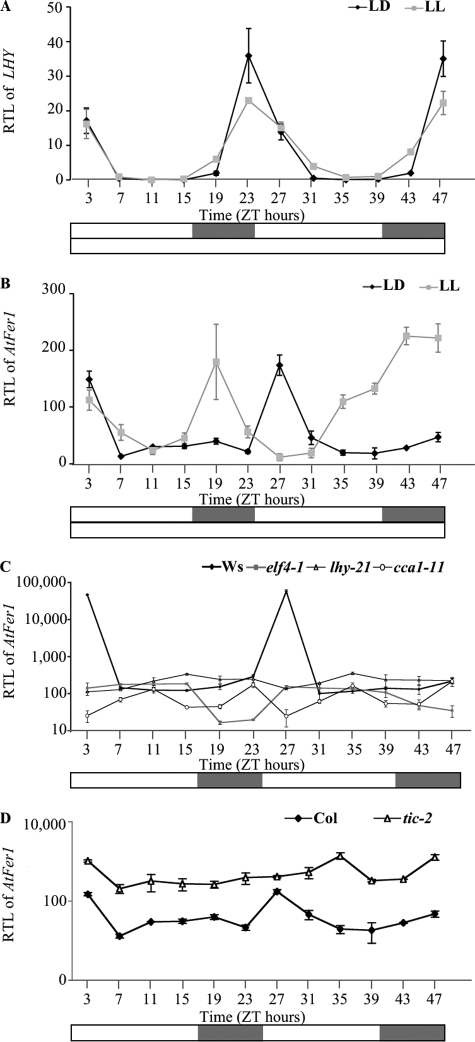
The TIC gene encodes a nuclear regulator of the circadian clock (12). Thus, a potential TIC-mediated regulation of AtFer1 expression through the circadian clock was investigated. Col plants were grown on soil in light/dark (LD) conditions then transferred in free running conditions i.e. in continuous light (LL) to discriminate between circadian and diurnal regulations. The persistence of rhythmic expression in such a condition is the signature of a clock-controlled gene. LHY (late elongated hypocotyl) (25, 26) was used as a control of circadian-controlled genes. LHY and AtFer1 mRNA accumulations were monitored every 4 h at the indicated Zeitgeber times for 48 h (Fig. 3, A and B). As expected, LHY mRNA accumulation presented exactly the same 24-h rhythm in LD and LL conditions (Fig. 3A). AtFer1 mRNA accumulation was rhythmic under LD conditions with a 24-h period (Fig. 3B) and peaked 3 h after dawn with a distinct phase of LHY, which peaked 1 h before dawn (Fig. 3A). Surprisingly, under free running conditions of continuous light, AtFer1 expression exhibited an 8-h phase advance compared with LD (Fig. 3B) and seemed to remain rhythmic at least during the first cycle. This suggested that AtFer1 expression could be controlled by the clock and responds to light changes with a high mRNA accumulation at dawn.
FIGURE 3.
Release experiments for analyzing the AtFer1 regulation. The plants were grown on soil under 16 h of light/8 h of dark (LD) cycles. Two-week-old plantlets were then transferred into LL or kept under LD cycles before harvesting leaves every 4 h for 2 days. RTLs were assayed by Q-PCR relatively to an internal EF1α control (At1g07930). A, RTL of LHY in Col plants under LD and LL conditions. B, RTL of AtFer1 in Col plants under LD and LL conditions. Black diamonds and gray squares represent, respectively, LD and LL (A and B). C, RTL of AtFer1 in Ws, elf4-1, lhy-21, and cca1-11 plants under LD conditions. Black diamonds, gray squares, open triangles, and open circles represent, respectively, Ws, elf4-1, lhy-21, and cca1-11. D, RTL of AtFer1 in Col and tic-2 plants under LD conditions. Black diamonds and open triangles represent, respectively, Col and tic-2. A–D, shown are the means ± S.D. (n = 3); open bars indicate light intervals, and closed bars indicate dark intervals; ZT, Zeitgeber time.
To confirm this hypothesis, mutants in the central oscillator of the circadian clock were used to analyze the AtFer1 expression in a clock breakdown background. The mutants impaired in the central oscillator used for the experiment were elf4-1 (27), lhy-21 (26), and cca1-11 (28), the insertion of the two latest being remapped (supplemental Fig. S2). ELF4 (early flowering 4) is one component of the evening loop of the oscillator (29), and the elf4-1 mutant showed a reduced capacity to anticipate dawn (30), making it relevant because AtFer1 expression is induced by dawn. LHY and CCA1 (circadian clock-associated 1) are components of the morning loop of the oscillator (29), allowing us to analyze AtFer1 response in both loops. The wild type parental line, Ws, displayed the same rhythmic AtFer1 expression as Col, whereas elf4-1, lhy-21 and cca1-11 did not lay out any rhythm in AtFer1 mRNA accumulation (Fig. 3C). Finally, we investigated the involvement of TIC, described as an upstream regulator of the central oscillator (12, 31), on AtFer1 expression for 48 h in plants grown in LD conditions (Fig. 3D). The AtFer1 mRNA level was higher in tic-2 compared with Col at each Zeitgeber time point (Fig. 3D), which is consistent with the up-regulation observed in the genetic screen. AtFer1 expression appeared arrhythmic in tic-2, a result expected for the effect of a component of the circadian clock on an output target (31, 32). Taken together, these results suggested that AtFer1 expression is regulated by the circadian clock through a TIC-dependent pathway.
Mutation in the TIME FOR COFFEE Gene Affects Iron Homeostasis in Arabidopsis
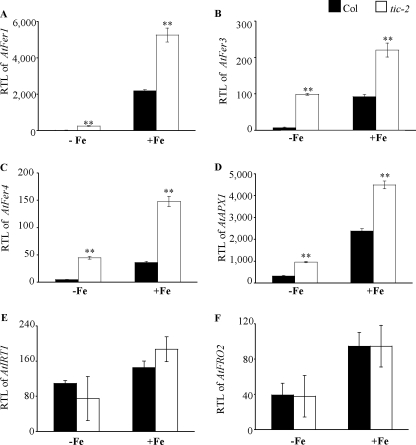
AtFer1 has been reported to be regulated by iron (9, 33); we thus investigated whether TIC is involved in the iron-dependent AtFer1 expression and more generally whether other iron-regulated genes could be regulated by TIC. AtFer1 expression was investigated in the tic-2 mutant in response to iron supply. The analysis was performed on leaves where AtFer1 is mainly expressed (8, 34). The expression of AtFer1 was analyzed on hydroponically grown plants with two iron supply conditions (iron deficiency and iron overload after deficiency). In iron-starved plants (−Fe) and in plants treated with iron citrate 500 μm for 5 h (+Fe), the expression of AtFer1 was higher in tic-2 than in Col (Fig. 4A). The fold of AtFer1 mRNA accumulation in response to iron was ∼150 in Col and ∼20 in tic-2. Thus, the AtFer1 mRNA abundance was still increased in response to iron excess in tic-2, and the smaller fold of increase in tic-2 seemed to be more related to a higher basal level in iron deficiency than to a defect in iron overload responsiveness. Under iron deficiency, AtFer1 expression was 17-fold higher in tic-2 than in Col. After iron overload, it was only 2.4-fold higher, suggesting that the TIC effect on AtFer1 expression was strongest under iron deficiency. This allowed us to conclude that TIC is necessary to repress AtFer1 gene expression under iron deficiency. Its impact appears to be the strongest under iron-deficient conditions at the molecular level, whereas the chlorotic phenotype was observable under iron sufficiency (Fig. 2A). Moreover, TIC is not involved in the iron responsiveness of AtFer1.
FIGURE 4.
Expression of iron-responsive genes in tic-2. Six-week-old plants, grown in hydroponic condition under iron sufficiency, were submitted to different iron treatments: iron deficiency during 1 week (−Fe) and iron overload for 5 h after 1 week of deficiency (+Fe). The leaves and roots were harvested separately, and RTL were measured by Q-PCR using PP2 (At1g13320), as a housekeeping gene (58). A, RTL of AtFer1 in leaves under iron deficiency and overload. B, RTL of AtFer3 in leaves under iron deficiency and overload. C, RTL of AtFer4 in leaves under iron deficiency and overload. D, RTL of AtAPX1 in leaves under iron deficiency and overload. E, RTL of AtIRT1 in roots under iron deficiency and overload. F, RTL of AtFRO2 in roots under iron deficiency and overload. Black and white bars represent, respectively, Col and tic-2; shown are the means ± S.D. (n = 3); **, p < 0.01 (A–D).
Like AtFer1, the AtFer3 and AtFer4 ferritin genes and the cytosolic ascorbate peroxidase AtAPX1 are also expressed in vegetative tissues, and their transcript abundance is increased in response to iron overload (8, 34, 35). We thus investigated whether the mutation in the TIC gene altered AtFer3, AtFer4, and AtAPX1 expression in leaves of plants grown under the same conditions as described above. The mRNA levels of these three genes were higher in tic-2 compared with Col in both iron nutrition conditions (Fig. 4, B–D). As observed for AtFer1, the fold of accumulation of these three transcripts in response to iron was lower because the level of mRNA accumulation was higher under iron starvation in tic-2. We next investigated whether the deregulation of these genes was directly related to the mutation in TIC, as for AtFer1, or indirectly resulting from a TIC-dependent increase of AtFer1 mRNA accumulation. Indeed, it was shown that the alteration of ferritin genes expression could impact the expression of other genes involved in iron homeostasis (8). For discriminating between these two hypotheses, a transgenic line overexpressing AtFer1 under the control of the 35S promoter (p35S::AtFer1) was used (supplemental Fig. S3A) and grown in the same conditions as above. The mRNA levels of AtFer3, AtFer4, and AtAPX1 were not affected in the overexpressing line, whatever the iron supply (supplemental Fig. S3, B–D). Thus, the overexpression of AtFer1 did not lead to the modulation of AtFer3, AtFer4, and AtAPX1 expression. This result indicated that TIC is necessary to keep AtFer3, AtFer4, and AtAPX1 gene expression at a low level under iron deficiency.
Because the effect of the tic-2 mutation was more obvious under iron deficiency and because tic-2 was chlorotic on soil, the expression of genes up-regulated in response to iron starvation was analyzed in the roots of wild type and tic-2 plants grown in the same conditions as described above. The AtIRT1 (iron(II) responsive transporter 1) (36) and AtFRO2 (ferric-chelate reductase oxidase 2) (37) genes were selected. In contrast to the iron excess-responsive genes analyzed, no significant difference in AtIRT1 (Fig. 4E) and AtFRO2 (Fig. 4F) mRNA accumulation was observed between wild type and tic-2 roots, regardless of the iron status. Thus, TIC has no impact on iron deficiency-regulated genes encoding the root iron uptake system, such as AtIRT1 and AtFRO2. By contrast, TIC represses the expression of iron overload-regulated genes, such as ferritins, under low iron conditions. Put together, these data showed that TIC is required for proper iron homeostasis in Arabidopsis.
TIC-mediated AtFer1 Regulation Is IDRS-independent
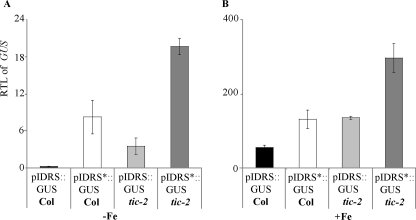
The data described above indicated that TIC acts in the repressive pathway leading to a low AtFer1 expression under iron deficiency (Fig. 4A). TIC regulates AtFer1 expression under repressive conditions through the circadian clock (Fig. 3). The IDRS box located in the AtFer1 promoter is a cis-acting element involved in the repression of AtFer1 transcription under iron deficiency (9). Consequently, we investigated whether the TIC-mediated AtFer1 regulation occurs through the IDRS. Two transgenic lines in Col background were used (9): At1400IDRS::GUS (named pIDRS::GUS Col) and At1400m*IDRS::GUS (named pIDRS*::GUS Col) carrying 1.4 kbp of the AtFer1 promoter region with, respectively, a wild type and a mutated IDRS sequence, fused to the GUS reporter gene. The two lines were crossed with tic-2, and homozygous lines for both loci were selected in the F2 progeny. They were named pIDRS::GUS tic-2 and pIDRS*::GUS tic-2. Seedlings of each line were grown in vitro for 10 days on iron-sufficient medium (50 μm iron-EDTA) and then transferred on an iron-depleted medium containing ferrozine 200 μm (−Fe) for 4 days. Afterward, half of the iron-starved plants were treated with 500 μm iron citrate for 5 h for iron overload treatment (+Fe). Both treatments were controlled by analyzing, respectively, the induction of AtIRT1 in iron-starved roots and of AtFer1 in iron-overloaded leaves (data not shown).
The transcript abundance of the GUS reporter gene was determined in leaves of the four transgenic lines (Fig. 5). As expected, the GUS mRNA abundance was higher in the pIDRS*::GUS Col and pIDRS::GUS tic-2 lines than in the pIDRS::GUS Col in both iron conditions (Fig. 5). On both iron nutrition conditions, the GUS mRNA accumulation was significantly higher in the pIDRS*::GUS tic-2 line than in the pIDRS*::GUS Col and pIDRS::GUS tic-2 lines. Moreover, the sum of the GUS RTL values for the pIDRS*::GUS Col and pIDRS::GUS tic-2 lines was almost equal to the RTL value for the pIDRS*::GUS tic-2 line. These data indicated that the tic-2 mutation and the IDRS mutated cis-element have independent and additive effects on the GUS mRNA abundance. Thus, it appears that AtFer1 is transcriptionally repressed by two independent pathways, one involving the cis-element IDRS and the other one involving the TIC gene.
FIGURE 5.
Involvement of TIC and the IDRS cis-element in the AtFer1 regulation. The tic-2 mutant was crossed with the transgenic lines pIDRS::GUS Col and pIDRS*::GUS Col to produce the lines pIDRS::GUS tic-2 and pIDRS*::GUS tic-2. The plants were grown in vitro under iron sufficiency during 10 days before being transferred for 4 days on iron-starved medium (−Fe). For the iron excess treatment, part of the iron-starved plants was sprayed with iron citrate 500 μm (+Fe). Leaves from plants grown on the different iron conditions were collected 5 h after the iron spray. The GUS RTL was measured in leaves by Q-PCR, using PP2 (At1g13320) as a housekeeping gene (58). A, RTL of the GUS reporter gene in iron deficiency. B, RTL of the GUS reporter gene in response to iron excess. Shown are the means ± S.D. with n = 3 (A and B).
TIC Requires LD Cycles and Light to Regulate AtFer1 Expression
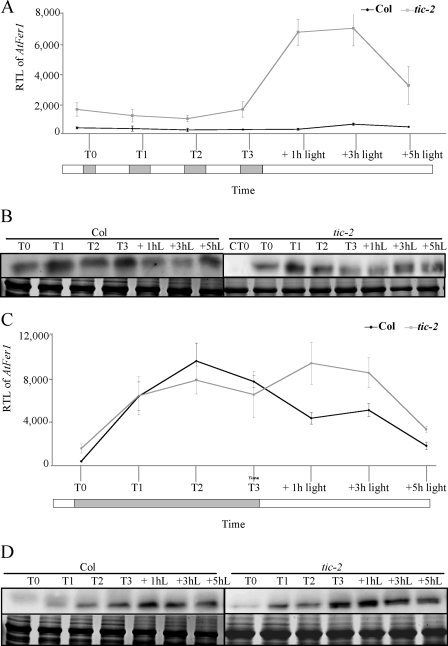
AtFer1 was reported to be up-regulated during dark-induced senescence (10). Because TIC seems to transduce the light signal to the central oscillator of the circadian clock and because AtFer1 expression responds to dawn or lighting by a huge increase in transcript accumulation (Fig. 3B), we wanted to determine whether the dark-induced AtFer1 expression depended on TIC, whether the effect of the tic-2 mutation was still effective in darkness, and finally whether the TIC-mediated regulation of AtFer1 expression required light. Col and tic-2 plants were grown on soil in LD conditions for 2 weeks. They were then maintained in LD conditions (Fig. 6, A and B) or transferred in continuous darkness for 3 days (Fig. 6, C and D). The samples were collected 30 min before dawn (T0) and then 24 h (T1), 48 h (T2), or 72 h (T3) later. After these 3 days, the light was switched on, and the samples were collected 1, 3, and 5 h after lighting.
FIGURE 6.
Dark-induced AtFer1 expression and response to a light return. Col and tic-2 plants were grown on soil under 16 h of light/8 h of dark cycles for 2 weeks. The 15th day, 30 min before lighting (T0), leaves of Col and tic-2 plants were collected afterward the remaining plants were kept in the same condition (LD) or transferred into continuous darkness. Triplicate samples were harvested every 24 h for 3 days (T1, T2, and T3). The third day, at the subjective dawn, the plants were turned back in light, and triplicate samples were harvested 1, 3, and 5 h after transfer. A, RTL of AtFer1 in LD. B, ferritin accumulation in LD; CT0, Col at T0. C, RTL of AtFer1 in continuous darkness. D, ferritin accumulation in continuous darkness. RTLs of AtFer1 were assayed by Q-PCR relatively to an internal EF1α control (At1g07930); shown are the means ± S.D. (n = 3); open and closed bars indicate, respectively, light and dark intervals (A and C). Ferritin accumulation was assayed by Western blot; 20 μg of total proteins, extracted from leaves, were loaded per lane, and immunodetection was performed using an anti-FER1 serum (16). The upper panel is the autoradiography (Ferr.), and the lower panel is the Coomassie Brilliant Blue staining used as a loading control (Coom.); hL, h of light treatment (B and D).
Under LD conditions, in tic-2, the accumulation of AtFer1 mRNA and of ferritin protein were systematically higher at dawn (Fig. 6, A and B), and AtFer1 mRNA was strongly overaccumulated in response to light (Fig. 6A). When plants were maintained in dark, AtFer1 mRNA and ferritin protein were highly accumulated in Col, as expected for a dark-induced gene. However, this accumulation was the same in tic-2 when compared with Col, suggesting that the TIC effect on AtFer1 expression was abolished by continuous darkness (Fig. 6C). The high accumulation of ferritin in response to continuous darkness also occurred at the protein level in tic-2 but slightly more quickly when compared with Col (Fig. 6D). When light was switched on after the 3 days of dark, AtFer1 transcript was slightly overaccumulated in tic-2, but not in Col, suggesting that light is required for TIC-repressive activity. Taken together, these results indicated that TIC is not involved in the dark-induced AtFer1 expression and that both LD cycles and light are required for the TIC repression of AtFer1 expression.
DISCUSSION
A LUC-based genetic screen using the AtFer1 iron-responsive promoter was initiated to identify regulators of iron homeostasis. This genetic screen led to the identification of TIME FOR COFFE as a new regulator of the AtFer1 gene expression. We investigated the function of TIC and of the central oscillator of the circadian clock on the expression of AtFer1 and of other iron overload-responsive genes. We showed that iron overload-expressed genes are regulated by TIC and by the central oscillator of the circadian clock. TIC represses their expression under low iron conditions, and its activity requires light and LD cycles. For AtFer1, this repression is independent of the previously characterized cis-acting element IDRS, known to be involved in AtFer1 repression. Finally, mutants in the TIC gene exhibited a chlorotic phenotype rescued by exogenous iron addition and are hypersensitive to iron during the early stages of development. These results showed that the regulation of iron homeostasis in plants is a major output of TIC-dependent and central oscillator-dependent signaling pathways.
TIC-, Circadian Clock-, and Light-mediated AtFer1 Regulation
The circadian clock generates endogenous 24-h rhythms and allows the anticipation of light and temperature changes in plants (31, 38). TIC was previously described as a nuclear regulator of the circadian clock in Arabidopsis and is specific to plants (12, 13). Because TIC is involved in a circadian network, we first investigated whether the TIC-mediated AtFer1 regulation occurs through a circadian mechanism. Our data showed that AtFer1 mRNA accumulation follows a 24-h rhythm in LD conditions (Fig. 3B). Surprisingly, in LL conditions, AtFer1 mRNA accumulation showed a 8-h phase advance compared with LD (19 h Zeitgeber time instead of 27 h Zeitgeber time) (Fig. 3B). It was previously shown that CAB expression peaked 6 h earlier in tic-1 than in the wild type and that the oscillator was arrested in this mutant after 19 h of light (13). The use of mutants impaired in the morning (lhy-21 and cca1-11) and in the evening (elf4-1) loops of the central oscillator confirmed that the TIC-mediated AtFer1 expression occurs through the clock, because AtFer1 mRNA accumulation was not rhythmic in these mutants under LD conditions (Fig. 3D). Put together, these results suggested that TIC regulates AtFer1 expression through a circadian pathway.
The rhythm of AtFer1 mRNA accumulation was lost in tic-2, and the transcript amount was systematically higher when compared with Col (Fig. 3D). This result showed that TIC represses AtFer1 expression in low iron condition and that the repressed state of AtFer1 is clock-regulated. The IDRS cis-element, located in the proximal region of the AtFer1 promoter, has already been shown to be involved in AtFer1 repression (9). A regulating pathway involving the trans-element TIC and the cis-element IDRS was therefore an attractive hypothesis to test. The analysis of Arabidopsis transgenic lines, carrying either a mutation in the TIC gene and/or in the IDRS, demonstrated that the effects of both mutations were additive and, thus, that the TIC-mediated AtFer1 regulation is independent of the IDRS (Fig. 5). It has been reported that, when plants carrying a pIDRS::GUS construct were subjected to a prolonged dark treatment, an increase of GUS activity was observed and that this phenomenon is IDRS-dependent (10). In our study, AtFer1 transcript accumulation and ferritin protein accumulation occurred similarly in Col and tic-2 exposed to prolonged darkness (Fig. 6, C and D), showing that the dark-induced expression of AtFer1 is independent of the TIC regulator, which is consistent with the IDRS independence of the TIC-mediated AtFer1 regulation (Fig. 5). Thus, AtFer1 expression is under the control of two independent repressive pathways: one involving the IDRS cis-element and controlling the dark-induced transcriptional activation and one involving TIC and controlling the circadian-dependent AtFer1 regulation.
AtFer1 expression is rhythmic, and the transcript amount peaks at dawn (Fig. 3B). Dawn is a time favorable for light-induced oxidative stress in leaves, and ferritins were shown to be involved in the protection against oxidative stress (8). Thus, it was consistent that the maximum level of AtFer1 mRNA abundance was observed at this time. We showed that the TIC-mediated AtFer1 regulation requires light and LD cycles (Fig. 6). TIC and ELF3 were reported to transduce the light signal to the central oscillator and to affect the circadian clock via the gating mechanism (12, 13, 39–41). The tic and the elf3 mutations seem to show an epistatic interaction, and phenotypes in the double mutant tic elf3 were additive or intermediate between the single mutants (12, 13). Thus, it should be interesting to analyze AtFer1 expression in elf3 and tic elf3 mutants to determine whether the rhythm and the dawn-induced transcript accumulation are affected. This will allow us to know whether ELF3, like TIC, is necessary for the regulation of AtFer1 expression, and whether they act redundantly.
TIC Controls Iron Homeostasis
Most of the studies regarding the regulation of ferritin expression focused on the transcriptional activation by iron (reviewed in Ref. 42). Thus, we investigated whether a link between the TIC-mediated regulation and the iron-signaling pathway exists, especially if TIC could affect other iron-responsive gene expression (Fig. 4). Indeed, AtFer3, AtFer4, and AtAPX1 expression was up-regulated in tic-2. The analysis of an AtFer1 overexpressing line clearly demonstrated that the increase of AtFer3, AtFer4, and AtAPX1 mRNA levels in tic-2 was not a secondary effect of the AtFer1 overexpression but resulted from the tic mutation itself. Consequently, besides AtFer1, TIC is also a repressor of AtFer3, AtFer4, and AtAPX1 expression. Interestingly, in contrast to AtFer1, which is derepressed by iron through the IDRS box (9), AtAPX1 is transcriptionally activated by iron (35). Thus, TIC acts as a repressor of both iron derepressed, i.e. AtFer1, and iron activated, i.e. AtAPX1, genes. By contrast, TIC did not affect the expression of iron deficiency-regulated genes such as AtIRT1 or AtFRO2, and that was independent of the iron supply conditions (Fig. 4, E and F). Interestingly, whereas ferritin overexpression increases root ferric reductase activity and leaf iron concentration in tobacco plants (43), the root iron uptake system was not activated in tic-2. This result suggested that overexpression of ferritin protein and a high ferritin gene expression arising from a TIC-mediated regulation defect did not lead to the same effect on the root iron uptake system.
Analysis of the tic-2 mutant showed that the mutation in the TIC gene led to phenotypes related to iron (Fig. 2). When grown on soil, tic-2 exhibited an iron chlorosis, but because the total iron content was not changed in the mutant, this suggested that tic-2 was plausibly affected in iron use or location at the cellular or at the subcellular level rather than in iron uptake or translocation from roots to shoots (Fig. 4) (44). When grown in vitro, tic-2 plantlets were affected in their early development on both iron-sufficient or excess media. Thus, TIC appears to be a central regulator of iron homeostasis in plants. This study made, for the first time, the link between the iron nutrition and the circadian clock and showed that the control of iron homeostasis appears to be a new output of the light/circadian clock pathways. In arable soil, iron is poorly available for plants (45), and iron starvation is commonly encountered. Iron is a limiting factor for plant productivity and biomass production (8, 46, 47). Thus, plants do probably not often experience iron overload. Therefore, in natural conditions where iron is poorly available, light, circadian clock, and TIC are likely to constitute the major signaling pathway of iron homeostasis regulation.
The next challenge will be to identify additional factors and components in this pathway, with special emphasis on the elements specific for the regulation of iron homeostasis. TIC is constitutively present among the circadian time (12), and AtFer1 expression is not all the time repressed by TIC. This raises the question of the regulation of TIC activity during circadian time. A putative P-loop motif, common to numbers of ATP/GTP-binding proteins, was identified in the TIC protein (12). It was suggested that this P-loop could bind to and be phosphorylated by a protein kinase (12). TIC activity could be regulated by (de)phosphorylation events (12), and interestingly, a PP2A-type phosphatase promotes an increase of AtFer1 mRNA level (33). Hence, TIC could regulate AtFer1 expression according to its phosphorylation state. The phosphorylated form could repress AtFer1 expression all the time, except at dawn when the unphosphorylated form would allow the increase of AtFer1 expression.
Recently, the Davis group screened for interactors of TIC and isolated the SNF1 stress-related kinase AKIN10 (48). AKIN10 is homologous to two members of the conserved energy sensor protein kinase family, SNF1 (sucrose nonfermenting 1) in yeast and AMPK (AMP-activated protein kinase) in mammals, and was proposed as a master metabolic sensor (49, 50). The SnRK1 (Snf1-related kinase) members AKIN10 and its homologue AKIN11 were shown to interact with SKP1/ASK1 (S phase kinase-associated protein 1/Arabidopsis SKP1-like 1) that mediates proteasomal binding of an ubiquitin ligase (51). This interaction is inhibited by PRL1 (pleiotropic response locus 1) that seemed to compete with SKP1 for binding to the C-terminal regulatory domain of AKIN10 and AKIN11. A recent genetic screen for identifying genes involved in the singlet oxygen signaling pathway (52) led to the isolation of a mutant in the PRL1 gene. Interestingly, the characterization of the prl1-5 mutant showed that the expression of AtFer1 and AtAPX1 was constitutively repressed in this mutant when compared with Col (53). This result suggested that PRL1 could be a positive regulator of the TIC activity. Thus, a regulatory network involving TIC, AKIN10, and PRL1 could be hypothesized for regulating iron homeostasis. TIC would be active when phosphorylated and would repress the AtFer and AtAPX1 genes. This active state would be (directly or indirectly) promoted by the AKIN10 kinase, and dawn favors the dephosphorylation of TIC, leading to the activation of the target gene expression. Because AKIN10 is negatively regulated by PRL1, AKIN10 could so constitutively activate TIC in prl1 mutants, leading to a lack of AtFer and AtAPX1 derepression at dawn and to a constitutive repression (53).
Input Signal Integration
Our study identified TIC as a major regulator of iron homeostasis in plants. Interestingly, two integrators of stress and energy signaling, AKIN10 (49) and PRL1 (53, 54), were recently shown to be connected to TIC and/or involved in the regulation of some iron-responsive gene expression. In response to energy deficit associated with stresses, the SnRK1 kinases seem to initiate genome-wide transcriptional changes that allow to restore homeostasis and to develop long term responses contributing to adaptation and preservation of growth and development. Regulatory mechanisms were evidenced to coordinate several inputs, such as light, circadian clock, or nutrient signals, into a complex signaling network in which only some insights have started to be understood (49, 55–57). Considering the essential functions of iron and iron-containing proteins in central metabolic processes such as photosynthesis, respiration, nitrogen, and sulfur assimilation (reviewed in Ref. 42), interplays regulating iron homeostasis should be assessed and elucidated to understand how the cellular energy signaling is fully integrated into whole plant adaptation and regulation of growth and development.
Acknowledgments
We thank Seth J. Davis (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for the gift of tic-1, tic-2, and elf4-1 seeds and Cécile Lambert for technical assistance in the map-based cloning. We thank the Salk Institute Genomic Analysis Laboratory (SIGNAL) for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the Nottingham Arabidopsis Stock Centre for providing seeds.
This work was supported by the Institut National de la Recherche Agronomique, the CNRS, and Action Concertée Incitative “Biologie Cellulaire Moléculaire et Structurale” Grant BCMS166 from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- IDRS
- iron-dependent regulatory sequence
- Q-PCR
- quantitative PCR
- RTL
- relative transcript level
- LUC
- luciferase
- LD
- light/dark
- LL
- continuous light
- EMS
- ethylmethane sulfonate.
REFERENCES
- 1.Cadenas E. (1989) Annu. Rev. Biochem. 58, 79–110 [DOI] [PubMed] [Google Scholar]
- 2.Andrews N. C. (2000) Nat. Rev. Genet. 1, 208–217 [DOI] [PubMed] [Google Scholar]
- 3.Andrews S. C., Robinson A. K., Rodriguez-Quinones F. (2003) FEMS Microbiology Reviews 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 4.Kim S. A., Guerinot M. L. (2007) FEBS Lett. 581, 2273–2280 [DOI] [PubMed] [Google Scholar]
- 5.Briat J. F., Curie C., Gaymard F. (2007) Curr. Opin. Plant. Biol. 10, 276–282 [DOI] [PubMed] [Google Scholar]
- 6.Briat J., Cellier F., Gaymard F. (2005) Ferritins and iron accumulation in plant tissues. In Iron Nutrition in Plants and Rhizospheric Microorganisms (Barton L., Abadia J. eds.), Kluwer Academic [Google Scholar]
- 7.Pilon M., Cohu C. M., Ravet K., Abdel-Ghany S. E., Gaymard F. (2009) Curr. Opin. Plant. Biol. 12, 347–357 [DOI] [PubMed] [Google Scholar]
- 8.Ravet K., Touraine B., Boucherez J., Briat J. F., Gaymard F., Cellier F. (2009) Plant J. 57, 400–412 [DOI] [PubMed] [Google Scholar]
- 9.Petit J. M., van Wuytswinkel O., Briat J. F., Lobréaux S. (2001) J. Biol. Chem. 276, 5584–5590 [DOI] [PubMed] [Google Scholar]
- 10.Tarantino D., Petit J. M., Lobréaux S., Briat J. F., Soave C., Murgia I. (2003) Planta 217, 709–716 [DOI] [PubMed] [Google Scholar]
- 11.Patton E. E., Willems A. R., Tyers M. (1998) Trends Genet. 14, 236–243 [DOI] [PubMed] [Google Scholar]
- 12.Ding Z., Millar A. J., Davis A. M., Davis S. J. (2007) Plant Cell 19, 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall A., Bastow R. M., Davis S. J., Hanano S., McWatters H. G., Hibberd V., Doyle M. R., Sung S., Halliday K. J., Amasino R. M., Millar A. J. (2003) Plant Cell 15, 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobréaux S., Thoiron S., Briat J. (1995) Plant J. 8, 443–449 [Google Scholar]
- 15.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 16.Liu Y. G., Mitsukawa N., Oosumi T., Whittier R. F. (1995) Plant J. 8, 457–463 [DOI] [PubMed] [Google Scholar]
- 17.Dellagi A., Rigault M., Segond D., Roux C., Kraepiel Y., Cellier F., Briat J. F., Gaymard F., Expert D. (2005) Plant J. 43, 262–272 [DOI] [PubMed] [Google Scholar]
- 18.Schaffner W., Weissmann C. (1973) Anal. Biochem. 56, 502–514 [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 20.MacKinney G. (1941) J. Biol. Chem. 140, 315–322 [Google Scholar]
- 21.Lobréaux S., Briat J. (1991) Biochem. J. 274, 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beinert H. (1978) Methods Enzymol. 54, 435–445 [DOI] [PubMed] [Google Scholar]
- 23.Briat J. F., Fobis-Loisy I., Grignon N., Lobréaux S., Pascal N., Savino G., Thoiron S., von Wiren N., Van Wuytswinkel O. (1995) Biol. Cell 84, 69–81 [Google Scholar]
- 24.Curie C., Briat J. F. (2003) Annu. Rev. Plant. Biol. 54, 183–206 [DOI] [PubMed] [Google Scholar]
- 25.Carré I. A., Kim J. Y. (2002) J. Exp. Bot. 53, 1551–1557 [DOI] [PubMed] [Google Scholar]
- 26.Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I. A., Coupland G. (1998) Cell 93, 1219–1229 [DOI] [PubMed] [Google Scholar]
- 27.Doyle M. R., Davis S. J., Bastow R. M., McWatters H. G., Kozma-Bognar L., Nagy F., Millar A. J., Amasino R. M. (2002) Nature 419, 74–77 [DOI] [PubMed] [Google Scholar]
- 28.Wang Z. Y., Tobin E. M. (1998) Cell 93, 1207–1217 [DOI] [PubMed] [Google Scholar]
- 29.Mas P. (2008) Trends Cell Biol. 18, 273–281 [DOI] [PubMed] [Google Scholar]
- 30.McWatters H. G., Kolmos E., Hall A., Doyle M. R., Amasino R. M., Gyula P., Nagy F., Millar A. J., Davis S. J. (2007) Plant Physiol. 144, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mas P. (2005) Int. J. Dev. Biol. 49, 491–500 [DOI] [PubMed] [Google Scholar]
- 32.Somers D. E., Webb A. A., Pearson M., Kay S. A. (1998) Development 125, 485–494 [DOI] [PubMed] [Google Scholar]
- 33.Arnaud N., Murgia I., Boucherez J., Briat J. F., Cellier F., Gaymard F. (2006) J. Biol. Chem. 281, 23579–23588 [DOI] [PubMed] [Google Scholar]
- 34.Petit J. M., Briat J. F., Lobréaux S. (2001) Biochem. J. 359, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fourcroy P., Vansuyt G., Kushnir S., Inze D., Briat J. F. (2004) Plant Physiol. 134, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vert G., Grotz N., Dedaldechamp F., Gaymard F., Guerinot M. L., Briat J. F., Curie C. (2002) Plant Cell 14, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson N. J., Procter C. M., Connolly E. L., Guerinot M. L. (1999) Nature 397, 694–697 [DOI] [PubMed] [Google Scholar]
- 38.Dunlap J. C. (1999) Cell 96, 271–290 [DOI] [PubMed] [Google Scholar]
- 39.McWatters H. G., Bastow R. M., Hall A., Millar A. J. (2000) Nature 408, 716–720 [DOI] [PubMed] [Google Scholar]
- 40.Covington M. F., Panda S., Liu X. L., Strayer C. A., Wagner D. R., Kay S. A. (2001) Plant Cell 13, 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X. L., Covington M. F., Fankhauser C., Chory J., Wagner D. R. (2001) Plant Cell 13, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briat J. F., Ravet K., Arnaud N., Duc C., Boucherez J., Touraine B., Cellier F., Gaymard F. (2009) Ann. Bot. (Lond) doi: mcp128 [pii]10.1093/aob/mcp128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Wuytswinkel O., Vansuyt G., Grignon N., Fourcroy P., Briat J. F. (1999) Plant J. 17, 93–97 [DOI] [PubMed] [Google Scholar]
- 44.Ravet K., Touraine B., Kim S. A., Cellier F., Thomine S., Guerinot M. L., Briat J.-F., Gaymard F. (2009) Molecular Plant doi:10.1093/mp/ssp041 [DOI] [PubMed] [Google Scholar]
- 45.Guerinot M. L., Yi Y. (1994) Plant Physiol. 104, 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blain S., Queguiner B., Armand L., Belviso S., Bombled B., Bopp L., Bowie A., Brunet C., Brussaard C., Carlotti F., Christaki U., Corbiere A., Durand I., Ebersbach F., Fuda J. L., Garcia N., Gerringa L., Griffiths B., Guigue C., Guillerm C., Jacquet S., Jeandel C., Laan P., Lefevre D., Lo Monaco C., Malits A., Mosseri J., Obernosterer I., Park Y. H., Picheral M., Pondaven P., Remenyi T., Sandroni V., Sarthou G., Savoye N., Scouarnec L., Souhaut M., Thuiller D., Timmermans K., Trull T., Uitz J., van Beek P., Veldhuis M., Vincent D., Viollier E., Vong L., Wagener T. (2007) Nature 446, 1070–1074 [DOI] [PubMed] [Google Scholar]
- 47.Cassar N., Bender M. L., Barnett B. A., Fan S., Moxim W. J., Levy H., Tilbrook B. (2007) Science 317, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 48.Sanchez Villarreal A., Davis S. J. (2009) TIME FOR COFFEE sets the circadian clock at dawn by integrating metabolic signals. In 20th International Conference on Arabidopsis Research, Edinburgh, UK [Google Scholar]
- 49.Baena-Gonzalez E., Rolland F., Thevelein J. M., Sheen J. (2007) Nature 448, 938–942 [DOI] [PubMed] [Google Scholar]
- 50.Baena-Gonzalez E., Sheen J. (2008) Trends Plant Sci. 13, 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farras R., Ferrando A., Jasik J., Kleinow T., Okresz L., Tiburcio A., Salchert K., del Pozo C., Schell J., Koncz C. (2001) EMBO J. 20, 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baruah A., Simkova K., Apel K., Laloi C. (2009) Plant Mol. Biol. 70, 547–563 [DOI] [PubMed] [Google Scholar]
- 53.Baruah A., Simkova K., Hincha D. K., Apel K., Laloi C. (2009) Plant J. doi: TPJ3935 [pii]10.1111/j.1365-313X.2009.03935.x [DOI] [PubMed] [Google Scholar]
- 54.Bhalerao R. P., Salchert K., Bako L., Okresz L., Szabados L., Muranaka T., Machida Y., Schell J., Koncz C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith A. M., Stitt M. (2007) Plant Cell Environ. 30, 1126–1149 [DOI] [PubMed] [Google Scholar]
- 56.Moore B., Zhou L., Rolland F., Hall Q., Cheng W. H., Liu Y. X., Hwang I., Jones T., Sheen J. (2003) Science 300, 332–336 [DOI] [PubMed] [Google Scholar]
- 57.Blasing O. E., Gibon Y., Gunther M., Hohne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W. R., Stitt M. (2005) Plant Cell 17, 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005) Plant Physiol. 139, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]