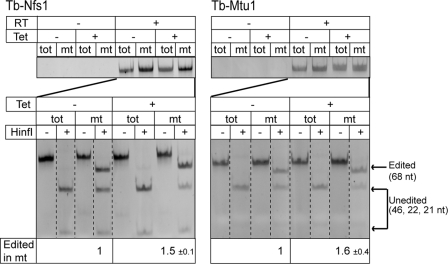
FIGURE 4.
Down-regulation of thio-modified tRNATrp stimulates C to U editing. Extent of tRNATrp editing in induced and uninduced Tb-Nfs1 and Tb-Mtu1 RNAi cell lines was quantified by an reverse transcription-PCR based assay as described before (12). The top panel shows that the cytosolic and mitochondrial RNA fractions that were used as templates for RT are free of DNA. The extent of RNA editing was analyzed by using a HinfI restriction digest (lower panel). RNA editing destroys a HinfI site that is only present in the cDNA derived from the unedited tRNATrp (14). Introduction of a synthetic HinfI at the 5′-end of the 5′ reverse transcription-PCR primer provides an internal control for the HinfI digestion. cDNA amplified from unedited tRNATrp contains two HinfI sites and, thus, will be digested into three fragments (46, 22, and 21 nt; unedited). The cDNA derived from edited tRNATrp contains the synthetic HinfI site only and will be digested into two fragments (68 and 21 nt; edited). Measuring the intensities of the diagnostic bands for non-edited (46 nt) and edited (68 nt) tRNATrp allows, after correcting for their different molecular mass, determination of the fraction of edited tRNATrp in T. brucei mitochondria. The mean of the increase, including standard error of tRNATrp editing in induced RNAi cells, is indicated. The restriction digests have been analyzed on a single gel for each RNA cell line, but the lanes have been rearranged electronically for clarity (dashed lines).