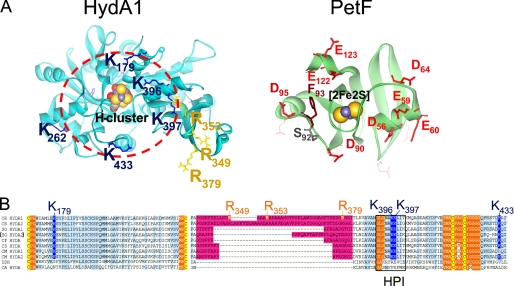
FIGURE 1.
Localization of exchange positions in structure models of HydA1 and PetF. Structures of HydA1 and PetF from C. reinhardtii (solid ribbon style; FeS-clusters in CPK style) were modeled using the crystal structures of CpI from C. pasteurianum (6) and PetF from Chlorella fusca (40) as archetypes. A, left side, detailed view on the assumed PetF-docking site, resolving the positively charged surface area of HydA1 with basic amino acid residues protruding from the brim of a surface recess leading to the [4Fe4S] subcluster of the active center. The depicted dark blue and yellow residues have been chosen for site-directed mutagenesis. The right side sketches the putative interaction surface on PetF. Residues that have been exchanged in this study are presented as red (acidic) or brown (aromatic) stick models. B, multiple sequence alignment of two parts of [FeFe] hydrogenase domains. A light blue background indicates a high level of sequence conservation. A pink background marks the large green alga-specific insertion. Cluster integrating cysteines are presented with a yellow background. Hydrophobic residues enclosing the H-cluster are indicated by an orange background. A black frame marks the sequence of the HPI. CR, C. reinhardtii (13, 16); SO, Scenedesmus obliquus (9); CF, C. fusca (10); CS, Chlorococcum submarinum (8); CM, Chlamydomonas moewusii (8); DDH, Desulfovibrio desulfuricans (41); CA, C. acetobutylicum (17).