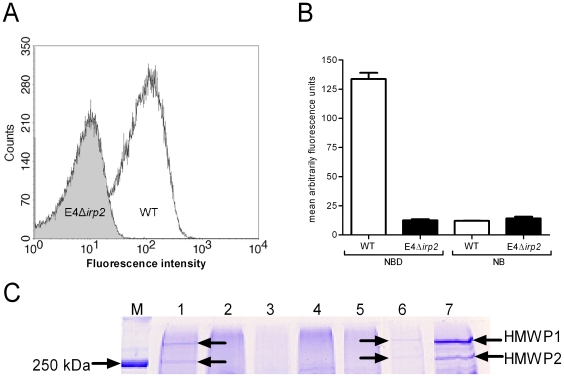
Figure 1. In the irp2 knockout strain the production of yersiniabactin is blocked.
Yersiniabactin production was tested using a GFP reporter assay with the knockout strain WA-CS irp1::Kanr containing the pCJG3.3N plasmid as the reporter strain to detect yersiniabactin in supernatants. The number of bacteria versus the measured arbitrary amount of fluorescence from 50,000 counted bacteria is shown in panel A. A) Supernatants from the irp2 knockout (E4Δirp2) cultures (grey) induced a lower amount of GFP compared to supernatants from the wild-type (WT) cultures (white). This indicates that production of yersiniabactin was blocked in the irp2 knockout. The experiments were performed in duplicate. B) The GFP reporter assay is dependent on yersiniabactin production, and production of yersiniabactin in the irp2 knockout is blocked. Yersiniabactin production is only seen in the wild-type (WT) cells cultured in iron-depleted medium (NBD), while the irp2 knockout (E4Δirp2) strain cultured in NBD and iron-containing medium (NB), as well as the wild-type strain grown in NB, did not produce yersiniabactin. Three different experiments were performed, with each sample analyzed in duplicate. C) The expression of HMWP1 and HMWP2 is disrupted by the insertion of a kanamycin resistance gene into irp2 using the Tagetron Knockout System. The irp2 knockout (E4Δirp2) strain was not able to produce HMWP1 and HMWP2 when cultured in NBD. M: marker; lane 1: wild-type strain E4; lane 2–5: irp2 gene knockouts created using the wild-type strain E4; lane 6 wild-type EHOS strain 03-702; lane 7: wild-type EHOS strain 03-819 served as HPI-positive control because the HMWP2 of this strain was confirmed by Edman degradation.